Abstract
Background
The incidence of liver disease is increasing in USA. Animal models had shown glutathione species in plasma reflects liver glutathione state and it could be a surrogate for the detection of hepatocellular carcinoma (HCC).
Methods
The present study aimed to translate methods to the human and to explore the role of glutathione/metabolic prints in the progression of liver dysfunction and in the detection of HCC. Treated plasma from healthy subjects (n = 20), patients with liver disease (ESLD, n = 99) and patients after transplantation (LTx, n = 7) were analyzed by GC- or LC/MS. Glutathione labeling profile was measured by isotopomer analyzes of 2H2O enriched plasma. Principal Component Analyzes (PCA) were used to determined metabolic prints.
Results
There was a significant difference in glutathione/metabolic profiles from patients with ESLD vs healthy subjects and patients after LTx. Similar significant differences were noted on patients with ESLD when stratified by the MELD score. PCA analyses showed myristic acid, citric acid, succinic acid, l-methionine, d-threitol, fumaric acid, pipecolic acid, isoleucine, hydroxy-butyrate and glycolic, steraric and hexanoic acids were discriminative metabolites for ESLD-HCC+ vs ESLD-HCC− subject status.
Conclusions
Glutathione species and metabolic prints defined liver disease severity and may serve as surrogate for the detection of HCC in patients with established cirrhosis.
Introduction
The incidence of end stage liver disease (ESLD) has increased in the last decade in the USA due mainly to a high incidence of HCV infection and an epidemic of obesity; the main risk factor for non-alcoholic steatohepatitis (NASH).1, 2 It is estimated that 4 million of Americans are infected with HCV and 1 out of 3 adults are overweight.3, 4 Non-invasive markers for liver disease progression are not well defined as they are the factors that predict the transformation from a compensated state of cirrhosis to a decompensated one.5, 6, 7 Furthermore, reliable biomarkers for early detection of hepatocellular carcinoma (HCC) in this high risk population are in need.8
Genetic prints on malignant tissue from patients with ESLD have shown diverse gene patterns likely due, at least in part to the heterogeneity in the mechanisms of liver injury and the variety of pathways for hepatocyte malignant transformation.9, 10, 11, 12, 13 In addition, there is no standardization in the method used to take and process tissue samples from tumor biopsies, or from tumor explants after surgery at different ischemic times during the operation. The integration of gene alterations to specific protein disturbances and metabolic patterns could provide a better understanding of the patient in whom disarrangements in liver physiology, portal blood flow, and malignant transformation occur at once.14, 15, 16, 17, 18 Our group has shown a linear correlation of glutathione species concentration in plasma with glutathione species concentration from liver tissue.19 Furthermore, an animal model with normal livers but tumor implants had shown disturbances of the glutathione species and changes on metabolic prints when compared to controls; changes in the glutathione species were also noted in an animal model with cirrhosis and HCC.20, 21, 22 The present study aims to translate methods from the animal model to the human, to determine changes in glutathione species and in the metabolic patterns of patients with progressive liver disease, and to determine differences in patients with ESLD with and without HCC. The present studies showed Glutathione species and metabolic prints defined liver disease severity and may serve as surrogate for the detection of HCC in patients with established cirrhosis.
Material and methods
Population
Three sets of subjects were enrolled on IRB approved protocols: healthy subjects (n = 20), patients with end stage liver disease (ESLD) and diverse liver injury (stratified by the MELD score, n = 99), and patients >30 days after liver transplantation (LTx) with normal graft function (n = 7). All subjects were approached during their visit to liver clinics, and protocols were presented and explained. Consented patients were scheduled for a second visit where clinical variables were captured, and blood and urine samples were obtained and processed. A subset of patients (healthy controls, n = 10; patients with diverse MELD score, n = 7 and patients after liver transplantation, n = 7) were enrolled for the labeling studies as described below. All studies were conducted according with the guidelines at Case Western Reserve University and Western IRB (Copernicus Group) under approved protocols at University Hospitals Case Medical Center (Cleveland) and Cancer Treatment Centers of America (Chicago).
The general dual protocol was aimed at i) measuring both the concentration and turnover of GSH-GSSG, and ophthalmate (from treated plasma and from 2H-enriched body water, respectively), and ii) evaluating the metabolome of the subjects over two Tiers of metabolites. In Tier 1, the concentration of glutathione species and 3 metabolites were measured (glucose, glycerol and lactate), while in Tier 2, the concentration of 78 metabolites were identified, respectively. An initial exploratory run of 21 metabolites was performed in healthy controls under fasted and fed conditions. To further assess liver physiology in healthy subjects without the need of getting a liver biopsy, pre-albumin and IGF-1 concentrations were measured by standard ELISA techniques (Sigma–Aldrich, St. Louis MO). All samples from patients with ESLD were taken in the fasted state in the morning hours under no sleep deprived conditions.23, 24
For the labeling studies, subjects were investigated at the Clinical Research Center (CWRU, Cleveland) and it involved: i) overnight fasting of the subject/patient, ii) insertion of a venous blood sampling catheter in a wrist vein, iii) collection of baseline blood and urine samples, iv) ingestion of glass-distilled 99% 2H2O (Sigma Aldrich, San Louis MO) in amount that enriched body water at 0.5% (250 ml/75 kg, ingested slowly over 1 h (the beginning of 2H2O ingestion sets the time at 0 h), v) blood sampling every hour from 1 to 8 h after starting 2H2O ingestion. Subjects/patients were asked to remain recumbent from 0 to 3 h to avoid vertigo or nausea and to assure better water distribution. Enrolled healthy subjects were asked for a second visit where the 2H2O was provided after the ingestion of a standardized 800 calorie meal.
Blood and urine sampling processing
Five mL of blood was withdrawn from patients using a vacutainer system and collected in a heparinized solution to be placed in a centrifuge at a temperature of 4 °C for 10 min. Plasma was carefully separated from RBC's and both aliquots were treated as described below; they were saved & labeled at −70 °C until mass spectrometry processing. 5–10 ml of urine were collected in pre-labeled sterile containers and saved at −70 °C. Urine samples were used in the identification of spectra peaks on unknown origin suspected to be from medications.
Dot connecting test
At the end of each study visit, every participant across the various study groups did a Dot-connecting test to assess grade of encephalopathy. The Dot connecting test consists of drawing a line connecting numbered dots already present on a sheet of paper, going from 1 to 15. The amount of time the subject spends performing the dot connecting test was recorded. It was administered in a set of six different formats (A–G) and the results were averaged and expressed in seconds.
Glutathione sp. concentration
Materials and reagents
General chemicals, as well as 2H2O (99%, glass distilled) and [13C2,15N-glycine]glutathione (M3 GSH) were from Sigma–Aldrich (St. Louis, MO). Homoglutathione was from Chem-Impex International (Wood Dale, IL). Ophthalmic acid was from Bachem (Torrance, CA). Acetonitrile was procured from Fisher Scientific (Pittsburgh, PA).
Glutathione species
Plasma concentration of glutathione species GSH:GSSG and OA were measured using LC-MS/MS methods validated in our laboratory.21 In brief, the protected plasma samples were spiked with homoglutathione as an internal standard. Iodoacetate treated samples were kept in the dark at room temperature for 45 min to allow completion of the reaction. To liberate the bound glutathione, 200 μl of DTT (100 mM, pH 10, in 10 mM ammonium bicarbonate) was added and allowed to react in the dark for 15 min at room temperature. To this solution, 200 μl of iodoacetonitrile [200 mM, pH 10, in 10 mM ammonium bicarbonate containing 3.12 μM homoglutathione (623 pmol/200 μl)] was added to convert the liberated glutathione to a cyanomethyl-thioether. After 30 min standing at room temperature in the dark, 1.5 ml of acetonitrile was added to precipitate the proteins. After completing all reactions, the sample was dried at 50 °C under air at 20 psi for 40 min and reconstituted in formic acid in water (0.1% vol:vol). This sample was injected into a liquid chromatograph (Agilent 1100, Agilent Technologies Inc., Palo Alto, CA) equipped with an API 4000 QTrap mass spectrometer (Applied Biosystems, Foster City, CA) operated under positive ionization mode. A Hypersil Gold C18 column (2.1 × 150 mm, 5 μm particle size; Thermo Electron Corp.) was used at room temperature. Mobile phase A was 0.15% formic acid in water-acetonitrile (99:1, vol:vol), and mobile phase B was 0.15% formic acid in water-acetonitrile (5:95 vol:vol). Using a gradient elution, the compounds were eluted at a flow rate of 0.2 ml/min. Analyst software (version 1.4.1; Applied Biosystems, Foster City, CA) was used for data registration and analysis. The MRM ion pairs monitored (precursor:product) were i) for carboxymethyl-GSH: 366.1:237.2, ii) for cyanomethyl-GSH derived from GSS-bound: 347.2:272.1, iii) for carboxymethyl-homoglutathione: 380.1:233.1, iv) for cyanomethyl-homoglutathione: 361.1:232.1, and v) for ophthalmate: 290.3:161.1. The GSH/GSSG-bound ratio was calculated using the formula [GSH]/[GSSG-bound2]. Concentrations were adjusted accordingly to internal controls.
Metabolic prints
Gas Chromatography-Mass Spectrometry (GC–MS) analyses were performed on an Agilent 6890 gas chromatograph interfaced to an Agilent 5973 mass spectrometer equipped with a Phenomenex ZB-5 MSi capillary column (30 m × 0.25 mmi.d., 0.25 μm film thicknesses). Injection volume was 1 μL in split less mode. Injector temperature was set at 250 °C and the transfer line at 275 °C. The carrier gas was helium at a constant flow rate of 1 mL/min. The GC oven temperature was initially kept at 60 °C for 1 min and increased at a rate of 10 °C/min to a final temperature of 325 °C held for 10 min. EI ion source temperature was set to 250 °C and the MS quadrupole temperature to 150 °C. Mass spectra were acquired in scan mode with a mass range of 45–800 m/z. The raw data was de-convoluted with the National Institute of Standards and Technology (NIST) Automated Mass Spectral Deconvolution and Identification Software (AMDIS). After spectral analysis and data processing of 113 signals, 78 signals could be identified in 80% of all samples. Identified signals were confirmed by the Case Core metabolomic library and the Fiehn library (Agilent Technologies Inc, Santa Clara, CA). For further quantification, the data was exported to the SpectConnect server (Massachusetts Institute of Technology, Cambridge, MA). The concentration of each metabolite was expressed as its relative peak area (divided by the area of the corresponding internal standard in the same chromatogram). All identified metabolic compounds in Tiers 1and 2 (3 and 78 metabolites, respectively) were further used for statistical analyses. For healthy controls, metabolic compound were measured and processed at T0 when compared to other groups and at T4 when healthy controls were compared between fasted and fed states.
Glutathione species labeling profile
When body water is 2H-enriched by ingestion of 2H2O, aminoacids become labeled by exchange reactions with the H of water. Once incorporated into proteins or peptides, the labeling of the aminoacids is stable. The measurement of the 2H-enrichment of proteins or peptides allows the calculation of their fractional rate of production. Since GSH is a peptide, its rate of synthesis in various organs can be accessed from the incorporation of 2H from 2H-enriched body water. Our laboratory had developed LC-MS/MS assays for the concentration and 2H-enrichment of GSH and GSSG. To prevent the oxidation of GSH, we immediately reacted the sample with iodoacetate or iodoacetonitrile to form the carboxymethyl-GS or cyanomethyl- GS thioether, respectively. These derivatives yield good MS–MS spectra. However, GSSG, which does not require protection, yields a very weak MS signal. To assay reliably GSH and GSSG under similar conditions, (i) we protect GSH with iodoacetate, then (ii) we reduce GSSG with dithiothreitol and protect the newly formed GSH with iodoacetonitrile. This one-pot preparation allows us to assay extant GSH and GSH derived from GSSG in the same LC-MS/MS run. Homo-GSH (glutamate-cysteine-alanine) is used twice as an internal standard as previously described.19
Treatment of samples and LC–MS/MS
The treatment of samples and the process to obtain the spectra were carried out as described previously.19 Analyst software (version 1.4.2; Applied Biosystems) was used for data registration. Mass spectra were acquired under three modes. First, the enhanced resolution mass spectra of the intact molecules was recorded. Second, multiple reaction monitoring (MRM) ion pairs were monitored to acquire data used to calculate isotopic enrichments from precursor 3-product pairs. The ion pairs monitored were i) for carboxymethyl-GSH: 366.1 → 237.1, 367.2 → 237.1, and 367.1 → 238.1; ii) for cyanomethyl-GSH derived from GSSR: 347.2 → 218.1, 348.2 → 218.1, and 348.2 → 219.1; iii) for carboxymethylhomoglutathione: 380.1 → 233.1; iv) for cyanomethyl-homoglutathione: 361.1 → 232.1; and v) for ophthalmate: 290.3 → 161.1, 291.3 → 161.1, and 291.3 → 162.1. Third, product ion spectra (MS/MS) were used to calculate the enrichments of fragments of the carboxymethyl-GSH derivative; from the precursor ions at mass-to charge ratio (m/z) 366.1 and 367.1, we monitor m/z 134 [cysteine (CYS) at collision energy 33], 237 [glycinyl (GLY)-CYS at collision energy 19], and 291 [glutamyl (GLU)-CYS at collision energy 21], with a mass width of 5 amu, from m/z 100 to 400. The 2H enrichment of plasma water was assayed by exchange with unlabeled acetone in alkaline medium, followed by extraction and GC–MS of deuterated acetone.
Calculations
The M1 2H enrichment of GSH derivatives was calculated from MRM data on the basis of the fragmentation patterns of the M and M1 parent ions of each derivative. The calculations for the carboxymethyl-GSH derivatives are based on the transitions of the M parent ion (366.1 → 236.1) and M1 parent ion (367.1 → 237.1 and 238.1). The M fraction is calculated from the average peak intensity of the 366.1 → 237.1 MRM pair. The M1 fraction is calculated as the sum of the average peak intensities of the two MRM pairs 367.1 → 237.1 and 367.1 → 238.1. The mole percent enrichment of the GSH derivative is calculated as M1/(M + M1). The contribution of M2 and higher mass isotopomers to the calculation is inconsequential. In experiments where M3 GSH was used, we measured the abundances of the M–M3 species. For the M2 isotopomer, we calculated the sum of the average peak intensities of three MRM pairs (368.1 → 237.1, 238.1, and 239.1). For the M3 isotopomer, we calculated the sum of average peak intensities of four MRM pairs (369.1 → 237.1, 238.1, 239.1, and 240.1). The M3 mole percent enrichment of the GSH derivative was calculated as M3/(M + M1 + M2 + M3. The above calculations use the average intensities of the peaks at retention time ±0.1 min. Data were processed using a Visual Basic script,1 which simplifies peak integration by opening the files sequentially, and then transferring the average intensity of the selected peaks to an Excel spreadsheet.
The M1 enrichment of the constitutive amino acids of GSH was calculated from the product ion spectra of the derivatives. Calculations used data from the most abundant product ions: m/z 134 (CYS), 237 (GLY-CYS), and 291 (GLU-CYS), monitored with a mass width of 5 amu. The enrichment of GLY was calculated from the difference in enrichment of GLY-CYS and CYS. The enrichment of GLU was calculated from the difference in enrichment of GLU-CYS and CYS. We fit the time course labeling data to a model for a single compartment using nonlinear regression implemented with the Origin statistical package (OriginLab, Northampton, MA). We used the Box-Lucas 1 model, which fits the data to the equation E(t) = Einf (1 − e−kt), where t is time and E(t) is the enrichment of the isotopomer at time t and Einf is the plateau enrichment. This equation describes the labeling of the pool with the tracer. The parameters to be estimated are the plateau enrichment and k, which is equal to flux/(pool size). We used the calculated 95% confidence intervals for the parameters as the criteria for determining difference when comparing the plots.
Statistical approach
Variables were entered in an Excel spreadsheet to be transferred to JMP statistical software (SAS Institute, Chicago V12). On continuous numerical non metabolic variables, probability of difference among groups was performed by ANOVA and between groups by a two-tailed t-test at a 0.05 level. Metabolites concentration data were normalized using log transformation. Difference between groups on categorical variables were tested by chi-square methods. Metabolic prints among groups were evaluated by principal component analyses as described below. Univariate analyses, t-test, and fold change analysis were performed to provide a preliminary overview about metabolites discriminating the groups. For the fold change analysis, a variable was considered as significant if the number was above the given threshold.
Principal component analyses
Multivariate analysis including Principal Component Analysis (PCA) and Partial Least Square Discriminant Analysis (PLS-DA) were conducted to investigate and visualize the pattern of metabolite differences among groups. PCA is an unsupervised method aiming to find the directions that best explain the variance in a data set. PLS-DA model used Variable Importance in the Projection (VIP) scores to identify the most influential metabolites. VIP scores higher than 0.8 are considered as meaningful. Metabolites with VIP score ≥ 1 were considered as significant important features to differentiate groups. Statistical analyses were performed using code developed in R (R Developmental Core Team http://www.R-project.org) and MetaboAnalyst 3.0 (Xia, J., Sinelnikov, I., Han, B., and Wishart, D.S. (2015) MetaboAnalyst 3.0 – making metabolomics more meaningful. Nucl. Acids Res. (http://dx.doi.org/10.1093/nar/gkv380).
Results
Main demographic variables of healthy subjects, patients with ESLD and patients after LTx are showed in Table 1. Healthy controls were significantly younger and with a lower MELD and Child Pugh Scores when compared with other groups of patients (p < 0.05). Patients with ESLD were divided into two subgroups according to their MELD score. Patients with ESLD either in the lower/higher MELD score group had similar proportion of HCC and similar distribution regarding the etiology for their liver disease.
Table 1.
Demographic variables of normal subjects, patients with different degrees of liver disease graded by MELD score and subjects after liver transplantation
| Group | Healthy | ESLD |
Post-LTx | p-Value* | |
|---|---|---|---|---|---|
| MELD < 14 | MELD ≥ 14 | ||||
| Subjects (n) | 20 | 79 | 20 | 7 | |
| Gender (M:F) | 11:9 | 43:36 | 15:5 | 4:3 | |
| Age (years M ± STDV) | 33.1 ± 8.7 | 55.9 ± 11.7 | 54.5 ± 15.1 | 55.4 ± 11.0 | <0.05 |
| MELD (M ± STDV) | 4.8 ± 1.5 | 8.9 ± 2.5 | 19.9 ± 6.2 | 5.7 ± 2.9 | <0.05 |
| Creatinine | 0.9 ± 0.04 | 0.97 ± 0.3 | 1.97 ± 1.2 | 1.6 ± 0.2 | |
| Bilirubin Total | 0.5 ± 0.2 | 1.12 ± 0.9 | 6.5 ± 11.2 | 1.1 ± 0.1 | |
| INR | 0.99 ± 0.0 | 1.13 ± 0.14 | 1.44 ± 0.5 | 1.1 ± 0.4 | |
| Child-Pugh Score | 5 ± 0 | 6.26 ± 1.3 | 8.7 ± 1.8 | 6.0 ± 1.7 | <0.05 |
| Ascites no controlled | 0 | 38 (48%) | 15 (75%) | 8 (28%) | |
| Encephalopathy (seconds) | 11.6 ± 5.3 | 14.8 ± 7.7 | 19.6 ± 11.6 | 19.4 ± 15.3 | |
| HCC (yes:no) | 0:20 (0%) | 13:66 (16%) | 3:18 (15%) | 0:7 (0%) | |
| Milan criteria | NA | 8 | 3 | NA | |
| Size | NA | 3.9 ± 1.7 | 2.5 ± 1.1 | NA | |
| Liver Disease | 0 | 79 | 20 | 7a | |
| Conge/metabolic (NASH) | NA | 16 | 3 | 2 | |
| Toxic/drug (alcohol) | NA | 13 | 6 | 1 | |
| Infectious (HBV/HCV) | NA | 41 | 10 | 3 | |
| Cholestatic | NA | 5 | 0 | 0 | |
| Other | NA | 4 | 1 | 1 | |
*p < 0.05 by t-test when compared to healthy controls.
Etiology of liver disease prior LTx.
Healthy subjects
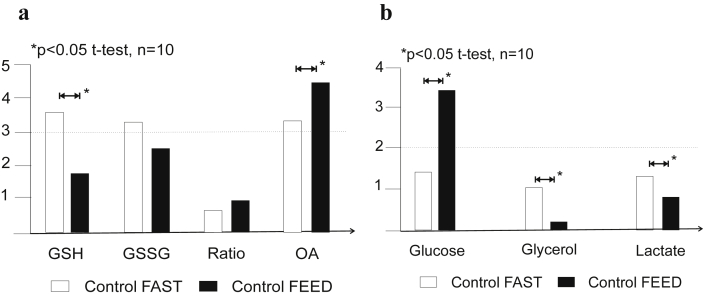
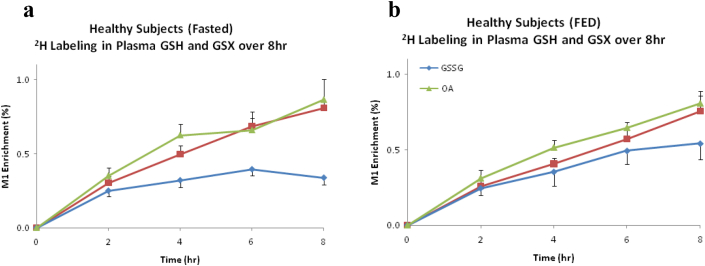
Twenty healthy subjects were included in the present studies; 11 males and 9 females with a mean age of 33.1 ± 8.7 years, and normal liver function test (not showed). Ten enrolled patients underwent labeling studies under fasted conditions and after a standardized 800 calorie meal. Healthy subjects showed higher concentration of GSH and a lower concentration of OA in the fasted state compared to the fed state (Fig. 1a, p < 0.05). Nevertheless, the turnover of GSH was similar (Fig. 2). Furthermore, healthy subjects differed significantly in the concentration of glucose, glycerol and lactate from the fasted state when compared to the fed state, as expected (Fig. 1b, p < 0.05). There were no significant differences in the glutathione species or Tier 1 metabolites when distributed by gender (male vs female, results no shown). In addition, there was not a statistical difference in the rate of labeling of GSH or OA between the fasted vs the feed state in the healthy subjects (Fig. 2). The results of their metabolic print on 21 compounds under fasted vs fed conditions are displayed on Table 2. The standardized 800 calorie meal was high in carbohydrates and proteins, and low in fats. The findings were consistent with our predictions.
Figure 1.
a. Glutathione species (GSH = glutathione reduced; GSSG = glutathione oxidized and bound; ratio = GSH/GSSG-bound2; OA = ophthalmic acid)) in healthy controls (n = 10) fasted for 8 h or after subjects were fed an 800 calories meal. Results are expressed as mean on relative concentration units. There was a statistical significant difference between the fast vs fed status in the relative concentrations of GSH and OA (t-test, p < 0.05). b. Tier 1 metabolites (glucose, glycerol and lactate) in healthy controls (n = 10) fasted for 8 h or after subjects were fed standardized 800 calories meal. There was a statistical significant difference between the fast status vs fed in the relative concentrations of glucose, glycerol and lactate (ANOVA followed by t-test, p < 0.05)
Figure 2.
Turn over of glutathione species. (GSH = reduced, GSSG = oxidized and bound and OA = ophthalmate) in healthy subjects (n = 10) by isotopomer analyses (M1) on2H2O enriched plasma. There was not a difference in the synthesis of glutathione sp. on healthy subjects a) Fasted for 8 h vs b) Fed a standardized 800 calorie meal (p > 0.05 by ANOVA)
Table 2.
Demographics and metabolite profile on 21 compounds of normal subjects. Subjects were fast for 8hr prior to the study vs fed a standardized 800calo meal just prior to the study. All studies were performed in the morning under no sleep deprivation and under standardized environment. Relative metabolomic data are expressed as ratios to an internal standard
| Healthy Controls |
p-Value* | ||
|---|---|---|---|
| FAST | FED | ||
| Gender (M:F) | 5:5 | 5:5 | |
| Age (years) | 44 ± 11 | 44 ± 11 | |
| Race | |||
| Caucasian | 6 | 6 | |
| African American | 2 | 2 | |
| Latino | 1 | 1 | |
| Body mass index | 25 ± 4 | 25 ± 4 | |
| Pre-albumin (g/dL) | 25 ± 8 | 23 ± 7 | |
| IGF-1 | 122 ± 47 | 123 ± 52 | |
| Co-morbidity | HTN (n = 1) | HTN (n = 1) | |
| Metabolite | |||
| Amino acids | |||
| Alanine | 0.26 ± 0.21 | 0.12 ± 0.04 | <0.05 |
| Glycine | 0.15 ± 0.12 | 0.17 ± 0.07 | |
| Isoleucine | 0.05 ± 0.03 | 0.09 ± 0.03 | |
| Lysine | 0.18 ± 0.016 | 0.12 ± 0.22 | |
| Proline | 0.15 ± 0.16 | 0.07 ± 0.04 | <0.05 |
| Serine | 0.08 ± 0.06 | 0.07 ± 0.03 | |
| Valine | 0.23 ± 0.28 | 0.16 ± 0.06 | |
| Threonine | 0.16 ± 0.12 | 0.13 ± 0.05 | |
| Carbohydrate | |||
| Glucose | 1.5 ± 1.7 | 3.47 ± 1.43 | <0.05 |
| Fructose | 0.02 ± 0.03 | 0.01 ± 0.01 | |
| Pyruvate | 0.01 ± 0.00 | 0.01 ± 0.00 | |
| Lactate | 1.23 ± 1.10 | 0.78 ± 0.33 | |
| α-Hydroxy-butyrate | 0.05 ± 0.06 | 0.03 ± 0.01 | |
| Amino-malonic acid | 0.02 ± 0.02 | 0.01 ± 0.01 | |
| Pyroglutamic acid | 0.13 ± 0.02 | 0.11 ± 0.06 | |
| Threonic acid | 0.01 ± 0.00 | 0.02 ± 0.00 | |
| Lipids | |||
| Cholesterol | 1.62 ± 0.83 | 2.44 ± 1.34 | |
| Glycerol | 0.09 ± 0.04 | 0.16 ± 0.08 | |
| Oleic acid | 0.50 ± 0.64 | 0.56 ± 0.51 | |
| Palmitate | 1.22 ± 1.06 | 0.98 ± 0.39 | |
| Stearic acid | 1.07 ± 0.76 | 1.12 ± 0.44 | |
*p < 0.05 by t-test.
Patients with end stage liver disease (ESLD)
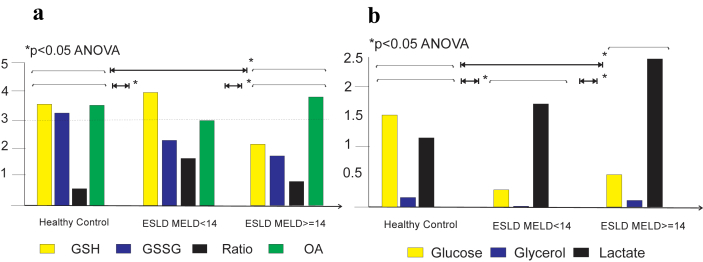
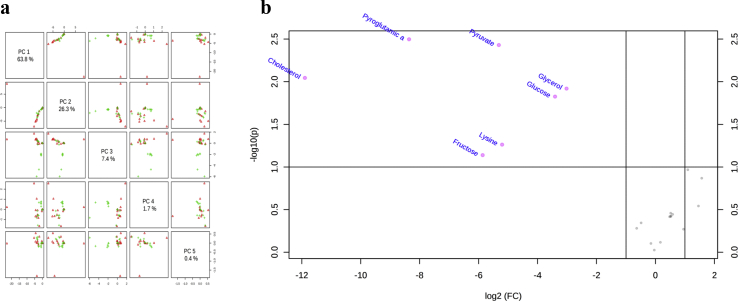
Glutathione species were significantly different in patients with ESLD when compared to healthy controls (Fig. 3a, p < 0.05). There was a decreased in the concentration of the reduced glutathione with change in the GSH/GSSG ratio. In addition, a significant difference in the Tier 1 metabolic print with marked decrease in glucose and glycerol and elevated concentration of lactated in patient with liver disease when compared to healthy subjects (Fig. 3b, p < 0.05). Alterations in all main metabolites (amino acids, carbohydrates and lipids) were also observed in patients with ESLD when compared with controls and those disturbances were more pronounced in patients with higher MELD score (>14). PCA showed pyro-glutamic acid, pyruvate, cholesterol, glycerol and glucose are significantly different in patients with ESLD when compared to healthy subjects. In addition, PLS-DA showed that lysine and fructose may also discriminate patients with ESLD from healthy subjects (Fig. 4).
Figure 3.
a. Glutathione species (GSH = glutathione reduced; GSSG = glutathione oxidized and bound; ratio = GSH/GSSG-bound2; OA = ophthalmic acid)) in fasted subjects from healthy controls (n = 20) and patients with end stage liver disease stratified by the MELD score (<14; n = 72) and ≥14; n = 27). Results are expressed as mean on relative concentration units. There was a statistical significant difference between the glutathione species print from patients with ESLD (either lower or higher MELD scores) compared to the print of healthy individuals (p < 0.05, by t-test). In addition there was a significant difference in the glutathione species print of patients with ESLD and lower vs higher MELD score (p < 0.05). b. Tier 1 metabolites (glucose, glycerol and lactate) in fasted subjects from healthy controls (n = 20) and form patients with end stage liver disease stratified by the MELD score (<14; n = 72) and ≥14; n = 27). There was a statistical significant difference between the Tier 1 metabolic print from patients with ESLD (either lower or higher MELD scores) compared to the print of healthy individuals (p < 0.05). In addition there was a significant difference in the Tier 1 metabolic print of patients with ESLD and lower vs higher MELD score (p < 0.05, by t-test)
Figure 4.
Principal Component Analyses (PCA) and Volcano Plot of metabolites in patients with ESLD. a. Tier 2 metabolites were measured in patients with end stage liver disease (ESLD) and healthy subjects. In was found pyro-glutamic acid, pyruvate, cholesterol and glucose significantly discriminate patients with ESLD and MELD ≥14 when compared to healthy controls. Results are displayed in b as a Volcano plot. In addition, PLS-DA, found lysine and fructose as additional discriminant metabolites between compared groups (VP scores no shown)
Patients with ESLD by etiology
A trend for some differences was observed in the glutathione species when patients with ESLD were grouped by their etiology of liver disease (Table 3). OA was noted to be significantly increased in patient with infectious causes (HCV/HBV). PCA and PLA-DA were not performed due to the low numbers of patients in some of the etiology groups.
Table 3.
Glutathione species of healthy subjects and patients with ESLD stratified by disease process categories: metabolic (NASH), toxic (alcohol related), infectious (HCV, HBV), cholestatic (autoimmune, PSC, PBC) and other (SBC, chronic rejection)
| Healthy | ESLD |
p-Value* | |||||
|---|---|---|---|---|---|---|---|
| Metab | Toxic | Infec | Choles | Other | |||
| Subjects (n) | 20 | 5 | 4 | 35 | 2 | 8 | |
| MELD | 4.3 ± 1.2 | 14 ± 5.3 | 17 ± 27.5 | 11.7 ± 4.8 | 11 ± 2.8 | 10.1 ± 2.3 | <0.05 |
| Child-Pugh | 5.0 ± 0.0 | 7.6 ± 2.3 | 8.5 ± 2.4 | 6.8 ± 1.5 | 6 ± 0 | 5.6 ± 0.7 | <0.05 |
| Encephalopathy (seconds) | 12.3 ± 5.2 | 13.8 ± 3.7 | 9.6 ± 2.7 | 16.7 ± 8.2 | 20.5 ± 12 | 29.2 ± 23 | <0.05 |
| GSH (nMol/ml) | 3.9 ± 1.6 | 2.5 ± 1.3 | 2.4 ± 1.2 | 3.5 ± 4.8 | 2.8 ± 0.5 | 2.6 ± 1.4 | |
| GSSG-bound | 3.6 ± 1.4 | 1.9 ± 1.0 | 1.8 ± 0.9 | 2.6 ± 2.8 | 1.5 ± 0.4 | 2.0 ± 0.8 | <0.05 |
| OA | 4.0 ± 4 | 6.4 ± 4.7 | 5.9 ± 3.1 | 8.9 ± 4.6 | 5.7 ± 4.4 | 4.9 ± 4.5 | <0.05 |
*p < 0.05 by t-test when compared to healthy controls.
Patients with ESLD and hepatocellular carcinoma (HCC)
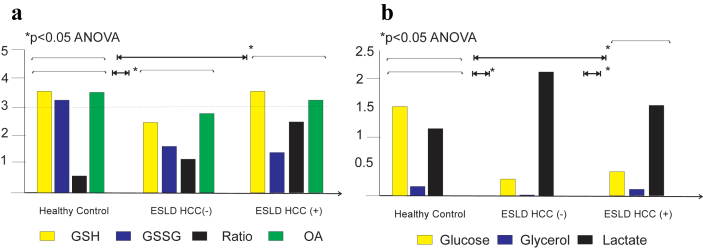
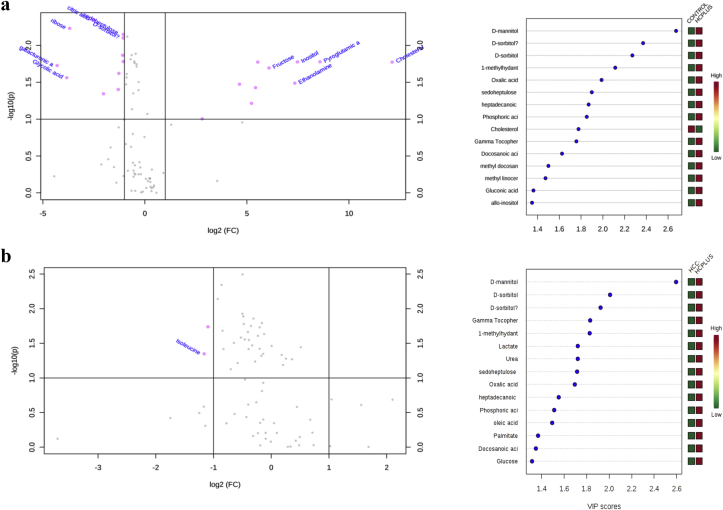
Patients with ESLD with and without malignancy showed a significant difference on their glutathione species profile when compared to controls (Fig. 5a, p < 0.05). Tier 1 metabolic print differed significantly in patients with ESLD by tumor status, ESLD & HCC− vs ESLC & HCC+ (Fig. 5b, p < 0.05). Tier 2 metabolites were measured in patients with ESLD and tumor vs healthy subjects. In was found, by PCA and PLS-DA cholesterol, pyroglutamic acid, inositol, ethanolamine, fructose, pyruvate, citric acid and lysine were discriminative metabolites. Additional metabolites displayed in the volcano plot included glycolic acid, galacturonic acid and ribose (Fig. 6). Tier 2 metabolites were measured in patients with ESLD-HCC+ vs ESLD-HCC−. PCA and PLS-da analyses showed myristic acid, citric acid, succinic acid, l-methionine, d-threitol, fumaric acid, pipecolic acid, isoleucine, glycolic acid, hydroy-butyrate, steraric acid and hexanoic acid were discriminative metabolites (Fig. 6). No additional metabolites were found in the volcano plot.
Figure 5.
a. Glutathione species (GSH = glutathione reduced; GSSG = glutathione oxidized and bound; ratio = GSH/GSSG-bound2; OA = ophthalmic acid)) in fasted subjects from three groups: healthy controls (n = 20), patients with end stage liver disease and no primary malignancy (HCC−, n = 83) and patients with end stage liver disease and primary malignancy (HCC+, n = 16). Results are expressed as mean on relative concentration units. There was a statistical significant difference between the glutathione species from patients with ESLD with or without malignancy compared to the print of healthy individuals (p < 0.05, by ANOVA followed by t-test). b. Tier 1 metabolites (glucose, glycerol and lactate) in fasted subjects from healthy controls (n = 20) and patients with ESLD by tumor status. There was a statistical significant difference between the Tier 1 metabolic print from patients with ESLD with or without malignancy compared to the print of healthy individuals (p < 0.05). Furthermore, there was a significant difference in the Tier 1 metabolic print of patients with ESLD and HCC− vs HCC+ (p < 0.05)
Figure 6.
Volcano Plot of metabolites by PCA & PLS-DA in patients with ESLD. a. Tier 2 metabolites were measured in patients with end stage liver disease (ESLD) and tumor (n = 16)) vs healthy subjects (n = 20). In was found cholesterol, pyroglutamic acid, inositol, ethanolamine, fructose, pyruvate, citric acid and lysine were discriminative metabolites. Additional metabolites displayed in the volcano plot included glycolic acid, galacturonic acid and ribose. Importance in projection (VIP) Scores for each metabolite found by PLS-DA are displayed. b. Tier 2 metabolites were measured in patients with ESLD-HCC+ (n = 16)) vs ESLD-HCC−(n = 68). It was found myristic acid, citric acid, succinic acid, l-methionine, d-threitol, fumaric acid, pipecolic acid, isoleucine, glycolic acid, hydroy-butirate, steraric acid and hexanoic acid were discriminative metabolites. No additional metabolites were found in the volcano plot. Importance in projection (VIP) Scores for each metabolite found by PLS-DA are displayed
Glutathione species labeling in patients with ESLD and after LTx
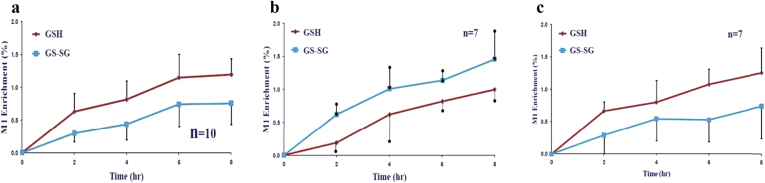
Patients with ESLD has a significant decreased in the labeling of GSH and an increased labeling of GSSG-bound when compared to healthy controls and patients after liver transplantation (p < 0.05). Patients after LTx has a similar profile as healthy controls (Fig. 7).
Figure 7.
Synthesis of glutathione sp. (GSH = reduced and GSSG = oxidized and bound) in fasted subjects from three groups: a) healthy controls (n = 10),b) patients with end stage liver disease (ESLD, n = 7) andc) patients after liver transplantation (LTx, n = 7). There was a statistical significant difference between the glutathione species labeling from patients with ESLD when compared to the glutathione synthesis from healthy individuals (p < 0.05 by ANOVA). The glutathione labeling from patients after liver transplantation were similar to the one from healthy patients
Discussion
The incidence of liver disease is increasing in the USA.1, 3, 4 ESLD is the most significant risk factor for the development of HCC, malignancy that has tripled its incidence in USA during the last decade.2, 25 Nevertheless, the detection of this malignancy on high risks population still relies on imaging modalities with up to 80% of cases being found at an advanced stage. The present study is aimed to determine metabolic prints and changes in the glutathione redox system in plasma from patients with ESLD. It was observed a significant different profile on patients with ESLD when stratified by MELD score when compared to the healthy subjects. Patients with ESLD had a significantly different glutathione species print and metabolic profile according to their tumor status. Patients with ESLD had a significantly different turn-over of glutathione species when compared to healthy subjects and to patients after liver transplantation.
We hypothesized that glutathione species print was a sensitive tool to detect oxidative stress of liver cells but not specific to the etiology of the injury. Nevertheless, the mechanism of the injury, as it differs according to the agent, may have a different metabolic print. Therefore, the measurement of glutathione species and the metabolic print of an individual may provide us with the severity of the insult and a metabolic print specific to the injury and to the state of liver disease and its progression. Patients with worsening liver dysfunction may have a different metabolic profile than a patient with progressive portal hypertension and both prints may add to the predictive value of the profile. In addition, the development of malignancy could not only aggravate the metabolic stress but also add a new profile. The present study showed that it is possible to i) differentiate patients with ESLD from healthy subjects based on the liver stress and their metabolic print, ii) to grade patients into low and high MELD scores (liver dysfunction), and iii) it may be possible to categorize subjects with ESLD by their tumor status. In addition, we may be able to follow the progress of patients by the etiology of their liver injury. Perhaps and instead of focusing on each metabolic path that may be altered in the individual or in the overall group, we would like to focus in ways that further define metabolic patterns of liver dysfunction and progression to ESLD and malignancy. Nonetheless, specific metabolic paths may serve as therapeutic targets or markers of response/failure to an intervention.
It was found that glutathione species and metabolic prints differentiated patients with low and high MELD score from healthy subjects. The MELD score predicts 90 days survival of patients with ESLD in the waiting list for LTx and predict postoperative complications after surgery of patients with liver disease.26, 27, 28, 29, 30 PCA and PLS-DA provided metabolic prints of patients with ESLD distributed by high or low MED scores. Thus changes in the glutathione species and metabolic patterns can measure progression of liver dysfunction, it could detect changes from a state of ESLD compensated to uncompensated one and perhaps predict survival of the patients in the LTx waiting list or graft function after transplantation. Furthermore, the metabolic changes noted during liver injury by a specific process, i.e. NASH, HCV had prints of common metabolites changes and prints of specific metabolites disturbances according to its etiology. The specific metabolic signature for an individual patient could be determined according to his/her etiology and disease progression. Early detection of a path for decompensation in a given etiology or for malignant transformation could alert the clinician for a timely intervention.31, 32 The number of patients required to validate such predictions are much larger and a matter of future studies.
Another aim of the present study was to translate methods from animal models where it was found glutathione species and metabolic disturbances were present in the early development of tumor cells; not only in normal livers but in livers with advanced fibrosis. Glutathione species and metabolic prints discriminate patients with ESLD and HCC from healthy patients, and more importantly they may discriminate subjects with tumors from patients with ESLD and no malignancy. Metabolic prints may prove to be a method for screening populations at high risk of developing HCC.33 The present study was not designed to correlate histology or tumor staging with a specific metabolic printing.34 Nevertheless, it would be a focus of another study to determine the metabolic print of primary liver tumors and their stage vs secondary tumors. In addition, we would like to determine if the grade of tumor differentiation could provide a specific metabolic printing that would predict and correlate with patient survival.18, 33
Glutathione species alterations were found to be a sensitive marker of liver stress imposed by any injury to the parenchymal cells and manifested in plasma on their reduced or oxidized forms. In addition, OA was noted to have similar sensitivity but also misses the specificity of the injury. The nature of the glutathione alterations was not approached in the present study neither their association with any specific etiology. Nevertheless, the labeling of glutathione reduced was significantly decreased while the labeling of glutathione oxidized was increased in patients with ESLD when compared to healthy subjects and to patients after liver transplantation with normal graft function. The higher disturbances were noted in patients with HCC and in patients with active viral infections. Even though the labeling of glutathione reduced was normalized after liver transplantation, the ratio of the reduced/oxidized glutathione forms and the concentration OA were increased in these patients when compared to healthy controls. This occurred even though they had normal graft function, perhaps reflecting an ongoing liver stress not obvious by routine liver tests.
The present studies should be taken in the light of some limitations. Standardization of procedures and MS techniques in a timely fashion are required if metabolic profiles are to be adopted for clinical use in the future. In addition, the further development of metabolic libraries will help to define and correlate specific metabolic prints with the specific state of liver dysfunction, portal hypertension, and tumor status for the individual. Since glutathione species and metabolic profiles are performed in processed plasma, it may prove to be a useful method for screening high risk populations for disease progression and perhaps by tumor status. Nonetheless, the number of patients presented in the present study are relatively low and values and profiles may vary as more studies are performed.
In conclusion, the present study has shown that glutathione species alterations and metabolic prints measured in plasma from patients with ESLD discriminated patients from healthy subjects, graded patients by the severity of their liver condition, and may categorize patients by the presence or absence of HCC. Further studies are needed to define the role of glutathione species and metabolic prints and their correlations with clinical variables to determine their predictive value and function as a screening tool.
Conflicts of interest
None declared.
Acknowledgment
This work was presented in part at the ASTS Philadelphia 2011, AHPBA/IHPBA Buenos Aires 2012 and ILTS Valencia 2012.
This work was supported by the Road Map grant initiative of NIDDK (5R33DK070291) and by Grant (1R01ES013925). Dr. RA Ibarra was supported by a Ruth L. Kirschstein National Research Service Award NIH/NIDDK (T32-DK007319).
Footnotes
This work was presented in part at the ASTS Philadelphia 2011, AHPBA/IHPBA Buenos Aires 2012 and AHPBA/IHPBA Sao Paulo 2016.
Contributor Information
Juan R. Sanabria, Email: sanabriaJ@marshall.edu, juan.sanabria@case.edu.
Rajan S. Kombu, Email: komburajan@gmail.com.
Guo-Fang Zhang, Email: gxz35@case.edu.
Yana Sandlers, Email: Yana.Sandlers@gmail.com.
Jizhou Ai, Email: Jizhou.Ai@ctca-hope.com.
Rafael A. Ibarra, Email: doctorrafaelibarra@gmail.com.
Rime Abbas, Email: d_reemy@yahoo.com.
Kush Goyal, Email: kgoyal@gmail.com.
Henri Brunengraber, Email: hxb8@case.edu.
References
- 1.Bunchorntavakul C., Reddy K.R. Diagnosis and management of overlap syndromes. Clin Liver Dis. 2015;19:81–97. doi: 10.1016/j.cld.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Global Burden of Disease Cancer C, Fitzmaurice C., Dicker D., Pain A., Hamavid H., Moradi-Lakeh M. The global burden of cancer 2013. JAMA Oncol. 2015;1:505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conzen K.D., Vachharajani N., Collins K.M., Anderson C.D., Lin Y., Wellen J.R. Morbid obesity in liver transplant recipients adversely affects longterm graft and patient survival in a single-institution analysis. HPB Off J Int Hepato Pancreato Biliary Assoc. 2015;17:251–257. doi: 10.1111/hpb.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts H.W., Utuama O.A., Klevens M., Teshale E., Hughes E., Jiles R. The contribution of viral hepatitis to the burden of chronic liver disease in the United States. Am J Gastroenterol. 2014;109:387–393. doi: 10.1038/ajg.2013.477. quiz 6, 94. [DOI] [PubMed] [Google Scholar]
- 5.Fox R.K. When to consider liver transplant during the management of chronic liver disease. Med Clin North Am. 2014;98:153–168. doi: 10.1016/j.mcna.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Johnson K.B., Campbell E.J., Chi H., Zheng H., King L.Y., Wu Y. Advanced disease, diuretic use, and marital status predict hospital admissions in an ambulatory cirrhosis cohort. Dig Dis Sci. 2014;59:174–182. doi: 10.1007/s10620-013-2832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patidar K.R., Thacker L.R., Wade J.B., Sterling R.K., Sanyal A.J., Siddiqui M.S. Covert hepatic encephalopathy is independently associated with poor survival and increased risk of hospitalization. Am J Gastroenterol. 2014;109:1757–1763. doi: 10.1038/ajg.2014.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong R.J., Wantuck J., Valenzuela A., Ahmed A., Bonham C., Gallo A. Primary surgical resection versus liver transplantation for transplant-eligible hepatocellular carcinoma patients. Dig Dis Sci. 2014;59:183–191. doi: 10.1007/s10620-013-2947-8. [DOI] [PubMed] [Google Scholar]
- 9.Bruix J. Usefulness of the molecular profile in the diagnosis, prognosis and treatment of hepatocellular carcinoma. Gastroenterol Hepatol. 2014;37(Suppl. 2):81–89. doi: 10.1016/S0210-5705(14)70074-3. [DOI] [PubMed] [Google Scholar]
- 10.Mah W.C., Thurnherr T., Chow P.K., Chung A.Y., Ooi L.L., Toh H.C. Methylation profiles reveal distinct subgroup of hepatocellular carcinoma patients with poor prognosis. PLoS One. 2014;9:e104158. doi: 10.1371/journal.pone.0104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishida N., Kudo M. Alteration of epigenetic profile in human hepatocellular carcinoma and its clinical implications. Liver Cancer. 2014;3:417–427. doi: 10.1159/000343860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Omar H., Lim C.R., Chao S., Lee M.M., Bong C.W., Ooi E.J. Blood gene signature for early hepatocellular carcinoma detection in patients with chronic hepatitis B. J Clin Gastroenterol. 2015;49:150–157. doi: 10.1097/MCG.0000000000000112. [DOI] [PubMed] [Google Scholar]
- 13.Yu F., Shen X.Y., Fan L., Yu Z.C. Genome-wide analysis of genetic variations assisted by ingenuity pathway analysis to comprehensively investigate potential genetic targets associated with the progression of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2014;18:2102–2108. [PubMed] [Google Scholar]
- 14.Armitage E.G., Barbas C. Metabolomics in cancer biomarker discovery: current trends and future perspectives. J Pharm Biomed Anal. 2014;87:1–11. doi: 10.1016/j.jpba.2013.08.041. [DOI] [PubMed] [Google Scholar]
- 15.Atzori L., Griffin J.L., Scherer P.E., Irminger-Finger I. Editorial for the directed issue: “Metabolic pathways in cancer”. Int J Biochem Cell Biol. 2011;43:948–949. doi: 10.1016/j.biocel.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 16.Baniasadi H., Gowda G.A., Gu H., Zeng A., Zhuang S., Skill N. Targeted metabolic profiling of hepatocellular carcinoma and hepatitis C using LC-MS/MS. Electrophoresis. 2013;34:2910–2917. doi: 10.1002/elps.201300029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barton R.H. A decade of advances in metabonomics. Expert Opin Drug Metab Toxicol. 2011;7:129–136. doi: 10.1517/17425255.2011.547473. [DOI] [PubMed] [Google Scholar]
- 18.Beger R.D. A review of applications of metabolomics in cancer. Metabolites. 2013;3:552–574. doi: 10.3390/metabo3030552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kombu R.S., Zhang G.F., Abbas R., Mieyal J.J., Anderson V.E., Kelleher J.K. Dynamics of glutathione and ophthalmate traced with 2H-enriched body water in rats and humans. Am J Physiol Endocrinol Metab. 2009;297:E260–E269. doi: 10.1152/ajpendo.00080.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abbas R., Kombu R.S., Ibarra R.A., Goyal K.K., Brunengraber H., Sanabria J.R. The dynamics of glutathione species and ophthalmate concentrations in plasma from the VX2 rabbit model of secondary liver tumors. HPB Surg. 2011;2011:709052. doi: 10.1155/2011/709052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ibarra R., Dazard J.E., Sandlers Y., Rehman F., Abbas R., Kombu R. Metabolomic analysis of liver tissue from the VX2 rabbit model of secondary liver tumors. HPB Surg. 2014;2014:310372. doi: 10.1155/2014/310372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ibarra R.A., Abbas R., Kombu R.S., Zhang G.F., Jacobs G., Lee Z. Disturbances in the glutathione/ophthalmate redox buffer system in the woodchuck model of hepatitis virus-induced hepatocellular carcinoma. HPB Surg. 2011;2011:789323. doi: 10.1155/2011/789323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dallmann R., Viola A.U., Tarokh L., Cajochen C., Brown S.A. The human circadian metabolome. Proc Natl Acad Sci U S A. 2012;109:2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies S.K., Ang J.E., Revell V.L., Holmes B., Mann A., Robertson F.P. Effect of sleep deprivation on the human metabolome. Proc Natl Acad Sci U S A. 2014;111:10761–10766. doi: 10.1073/pnas.1402663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.(NCI) NCI . 2015. SEER fast stats.wwwncigov [Google Scholar]
- 26.Borzio M., Dionigi E., Rossini A., Toldi A., Francica G., Fornari F. Trend of improving prognosis of hepatocellular carcinoma in clinical practice: an Italian in-field experience. Dig Dis Sci. 2015;60:1465–1473. doi: 10.1007/s10620-014-3427-5. [DOI] [PubMed] [Google Scholar]
- 27.Donadon M., Costa G., Cimino M., Procopio F., Fabbro D.D., Palmisano A. Safe hepatectomy selection criteria for hepatocellular carcinoma patients: a validation of 336 consecutive hepatectomies. The BILCHE score. World J Surg. 2015;39:237–243. doi: 10.1007/s00268-014-2786-6. [DOI] [PubMed] [Google Scholar]
- 28.Nobel Y.R., Goldberg D.S. Variable use of model for end-stage liver disease exception points in patients with neuroendocrine tumors metastatic to the liver and its impact on patient outcomes. Transplantation. 2015;99:2341–2346. doi: 10.1097/TP.0000000000000723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vicco M.H., Rodeles L., Ferini F., Long A.K., Musacchio H.M. In-hospital mortality risk factors in patients with ascites due to cirrhosis. Rev Assoc Med Bras. 2015;61:35–39. doi: 10.1590/1806-9282.61.01.035. [DOI] [PubMed] [Google Scholar]
- 30.Zielsdorf S.M., Kubasiak J.C., Janssen I., Myers J.A., Luu M.B. A NSQIP analysis of MELD and perioperative outcomes in general surgery. Am Surg. 2015;81:755–759. [PubMed] [Google Scholar]
- 31.Beyoglu D., Idle J.R. The metabolomic window into hepatobiliary disease. J Hepatol. 2013;59:842–858. doi: 10.1016/j.jhep.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beyoglu D., Imbeaud S., Maurhofer O., Bioulac-Sage P., Zucman-Rossi J., Dufour J.F. Tissue metabolomics of hepatocellular carcinoma: tumor energy metabolism and the role of transcriptomic classification. Hepatology. 2013;58:229–238. doi: 10.1002/hep.26350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aboud O.A., Weiss R.H. New opportunities from the cancer metabolome. Clin Chem. 2013;59:138–146. doi: 10.1373/clinchem.2012.184598. [DOI] [PubMed] [Google Scholar]
- 34.Brown M.V., McDunn J.E., Gunst P.R., Smith E.M., Milburn M.V., Troyer D.A. Cancer detection and biopsy classification using concurrent histopathological and metabolomic analysis of core biopsies. Genome Med. 2012;4:33. doi: 10.1186/gm332. [DOI] [PMC free article] [PubMed] [Google Scholar]