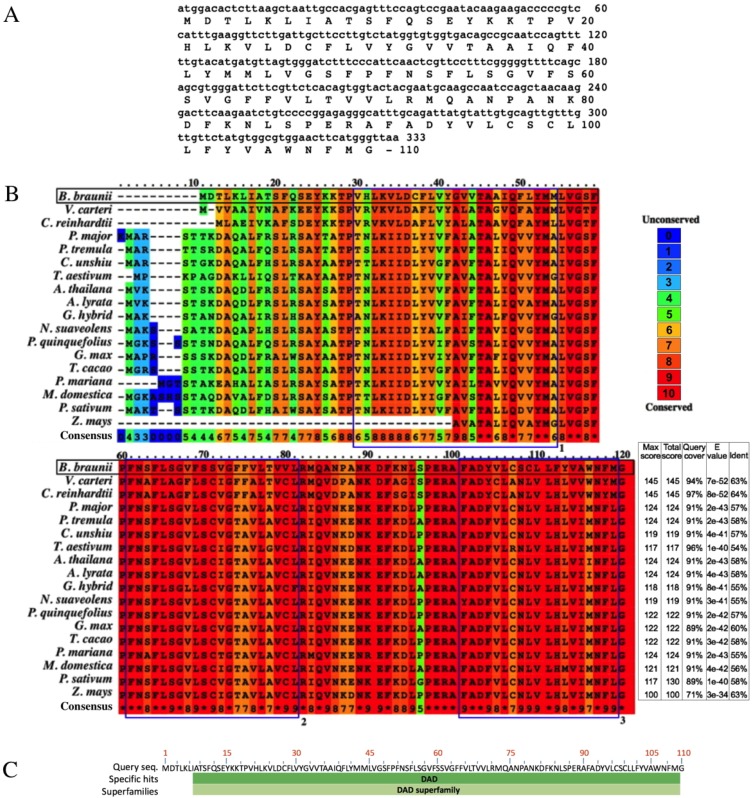
Figure 4. Nucleotide and amino acid sequences of DAD1, and protein alignment analysis.
Nucleotide and amino acid sequences of the B. braunii DAD1 (A), Alignment of DAD1 protein sequences from (B) Volvox carteri (XP_002956617), Chlamydomonas reinhardtii (XP_001699953), Plantago major (AM_156929), Populus tremula (AF086839), Citrus unshiu (AB011798), Triticum aestivum (GU564293), Arabidopsis thaliana (NM_102954), Arabidopsis lyrata (XP_002893712), Gladiolus hybrid (BAD11075), Nicotiana suaveolens (AB058922), Panax quinquefolius (EU274651), Glycine max (NP_001236226), Theobroma cacao (EOY25213.1), Picea mariana (AF_051247), Malus domestica (NP_001280846), Pisum sativum (AAC77357), and Zea mays (AF055909). Alignment was generated using PRALINE multiple sequence program. The histogram depicts level of conservation for each amino acid. Blue boxes labeled 1, 2, and 3 indicate the putative transmembrane domains. E-values and % of identity in comparison with B. braunii DAD1 are shown at the end of the sequences. The DAD superfamily domain (pfam02109) map was generated using blastp (C).