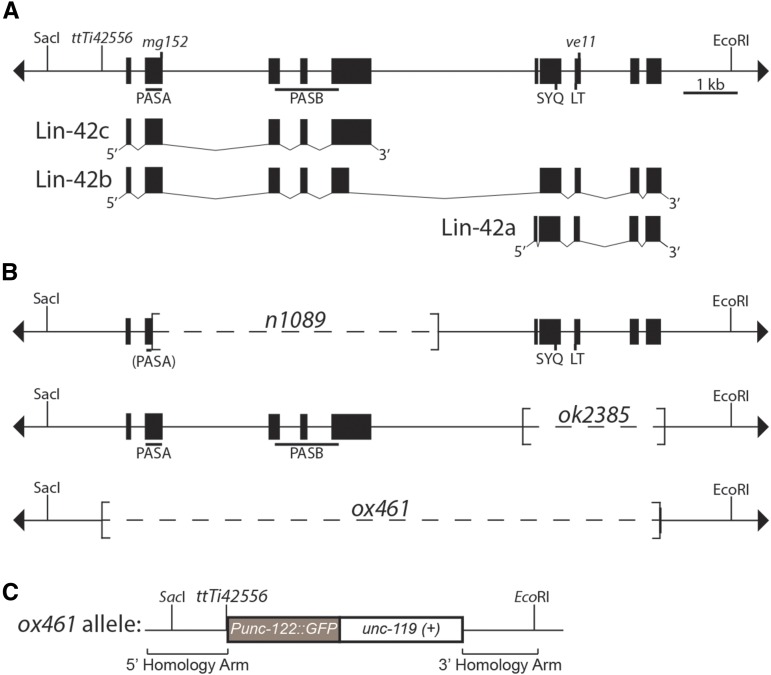
Figure 1.
lin-42(ox461) deletes the lin-42 coding region. (A) lin-42 genomic locus and transcription units. The line with terminal arrowheads represents genomic DNA of the lin-42 locus drawn with 5′ to the left, which is inverted from WormBase. lin-42 alleles and the Mos1 insertion (ttTi42556) are indicated above the line. SacI and EcoRI sites are included for reference with (B) and (C), but not all recognition sites for these enzymes are shown. The lin-42 locus produces three transcription units diagrammed below, with filled boxes representing exons: lin-42a, lin-42b, and lin-42c. lin-42a and lin-42c are nonoverlapping and expressed from distinct promoters (Tennessen et al. 2006). Note that the lin-42 nomenclature used here conforms to that adopted by WormBase and differs from that of pre-2014 publications from the Rougvie laboratory (e.g., Tennessen et al. 2006, 2010). (B) lin-42 deletion alleles. The extent of each deletion is noted in brackets. The lin-42(n1089) PASA domain is in parentheses since the majority of the domain is deleted. (C) The lin-42(ox461) allele deletes the lin-42 coding region and replaces it with Punc-122:gfp and C. briggsae unc-119(+). The fragments used as repair templates in creation of the deletion allele are indicated. See Materials and Methods for details.