Figure 5.
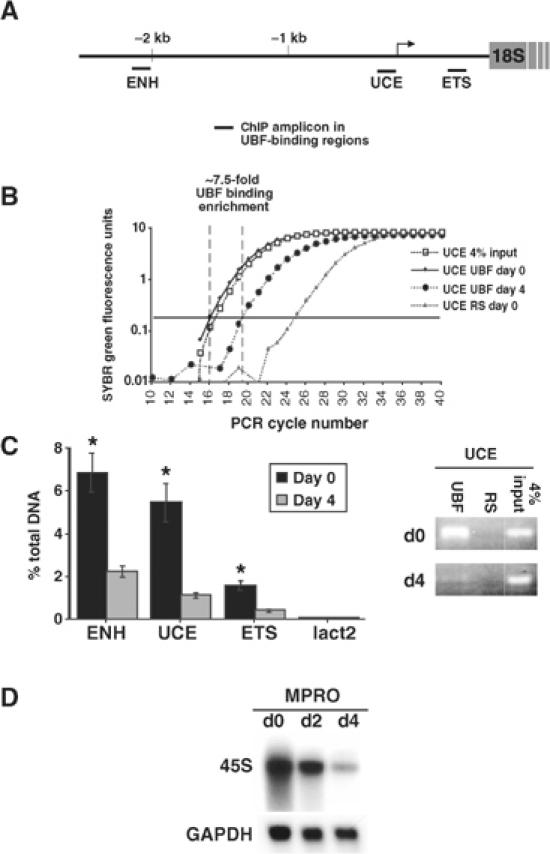
Regulation of UBF binding to the rDNA gene during granulocyte differentiation. MPRO cells were differentiated for 4 days and UBF binding to the rDNA gene was determined by ChIP assays using qRT-PCR. (A) Three regions of the murine rDNA gene were examined: the 5′ enhancer region (ENH); the upstream control element (UCE); and the 5′ external transcribed spacer (ETS). (B) Representative qRT-PCR amplification curves as displayed by ABI Prism 7000 are shown for the UCE amplicon. RS: control ChIP with preimmune rabbit sera. The arbitrary amplification threshold is depicted as the horizontal bar running across the graph. (C) Data presented as % of DNA site immunoprecipitated with anti-UBF compared to that present in total input DNA (based on 4% input; calculated as described in Frank et al, 2001). A site within the Lactoferrin promoter (lact2) was used as a negative control. Results are the mean±s.e.m. from three independent experiments. *P<0.05 compared to day-4 ChIPs. The representative ethidium bromide gel shows the amount of UCE product present after 19 cycles (within the exponential phase) of PCR of ChIP samples. (D) Cells were assayed for 45S rRNA expression by Northern blot (GAPDH control).
