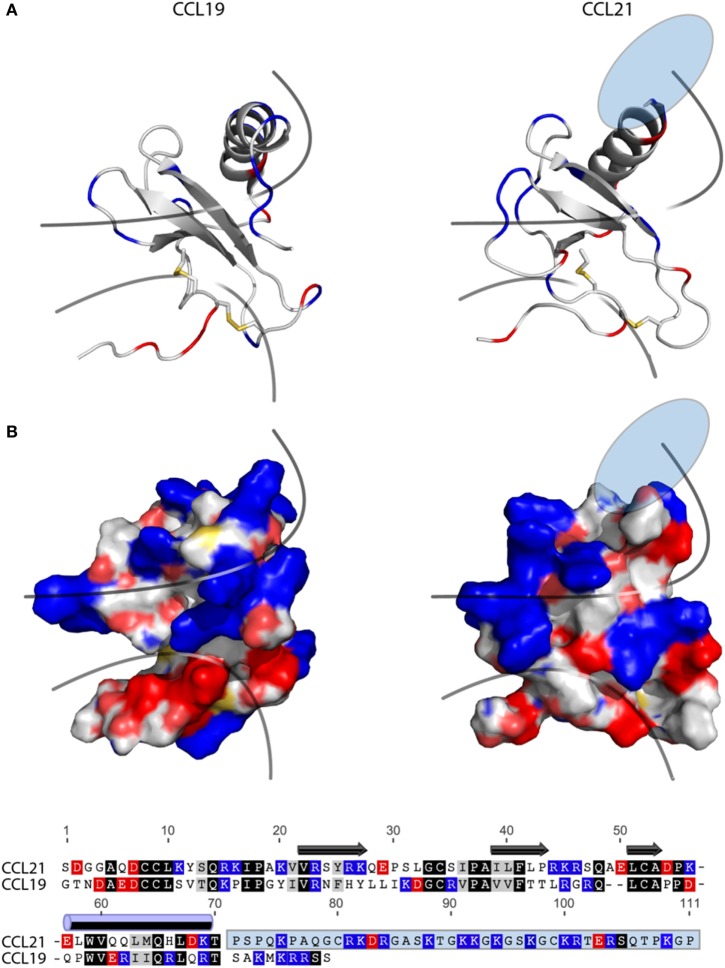
Figure 6.
Structure and alignment of CCL19 and CCL21. (A) NMR solution structure of CCL21 (PDB reference 2L4N) and CCL19 (PDB reference 2MP1). Cartoon structures are aligned in PyMOL to demonstrate the secondary structures likely to be found in both proteins (top). Exposed charges of either structure are shown on the surface presentation (bottom) with positive charges in blue and negative charges in red. Pale blue and pale red represents surface-exposed nitrogen- and oxygen atoms, respectively. The lower line divides the chemokine core domain from the highly flexible N-terminus, whereas the upper line delimits the regions that most closely match the mark of a GAG-binding domain, i.e., a dense cluster of exposed positive charges. The light blue ellipse on CCL21 represents the large unsolved C-terminal tail, of which most is also removed in tailless-CCL21. (B) Amino acid sequence alignment of CCL19 and CCL21 using the MAFFT multiple-aligner plug-in of Geneious Pro 6.1.7 software (Biomatters Ltd., Auckland, New Zealand). The secondary structures are shown with symbols, using arrows for beta-strands and a cylinder for the alpha-helix. Identical residues are black, similar residues are gray, and positively and negatively charged residues are blue and red, respectively. The unsolved C-terminus of CCL21 is outlined in a light blue box.