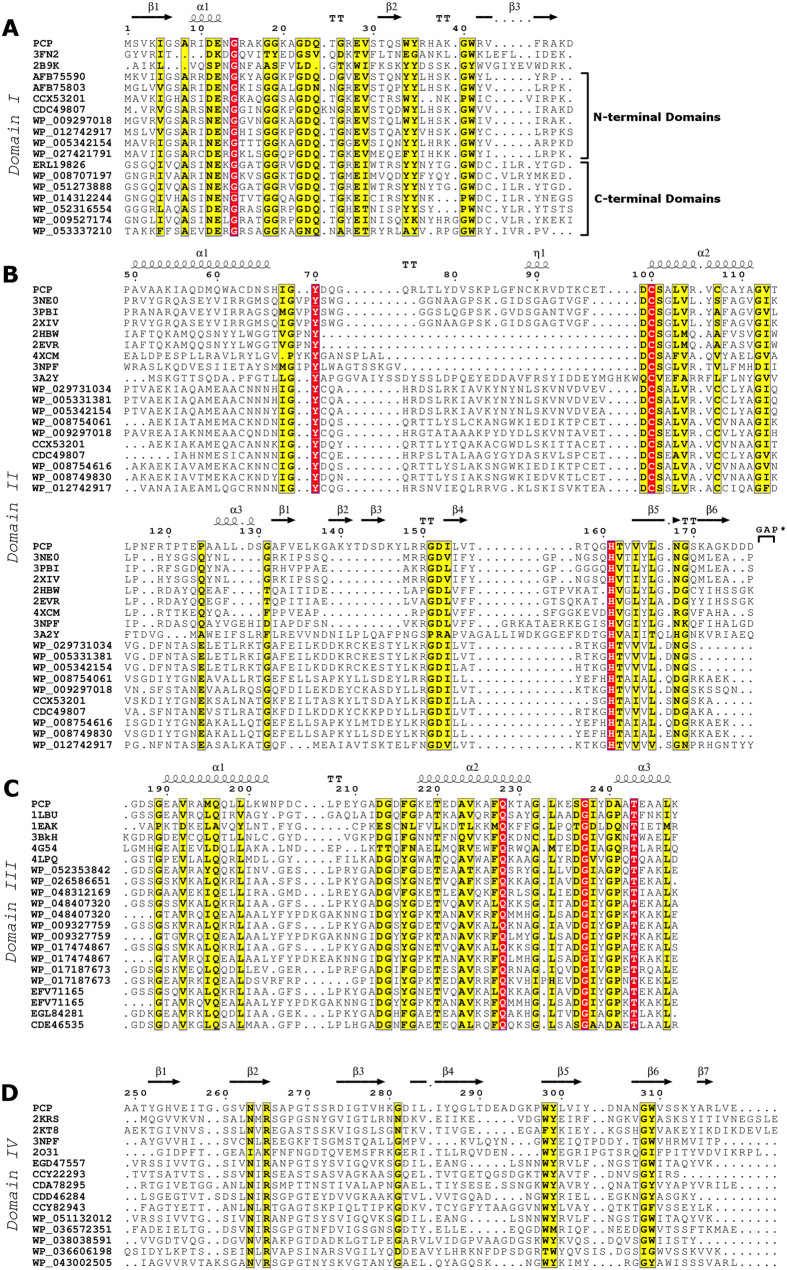
Figure 3. Sequence analysis of PCP domains. Secondary structures and residue numbering correspond to the PCP homology model and sequence, respectively.
(A) Domain 1 sequence alignment to homologous sequences and PDB structures*. Homologous sequences found at the C-terminus or N-terminus of catalytic domains are grouped and identified in the insert. (B) Domain 2 sequence alignment to homologous sequences and PDB structures*. Conserved catalytic Cys100 and His161 residues are highlighted in red. At the end of this domain there are 8 residues missing referred to as GAP: 178PVTDALKR185. (C) Domain 3 sequence alignment to homologous sequences and PDB structures*. Conserved PBD signature sequence repeat can be seen at position Asp213-Thr230 and Asp236-Thr243 (in PCP residue 236 is a Ser). (D) Domain 4 sequence alignment to homologous sequences and PDB structures*. *All PDB structures are identified by their four-character accession codes (ex. 3FN2, 3NE0, 1LBU, 2KRS etc). This figure was produced using the ESPript online server (http://espript.ibcp.fr)89.