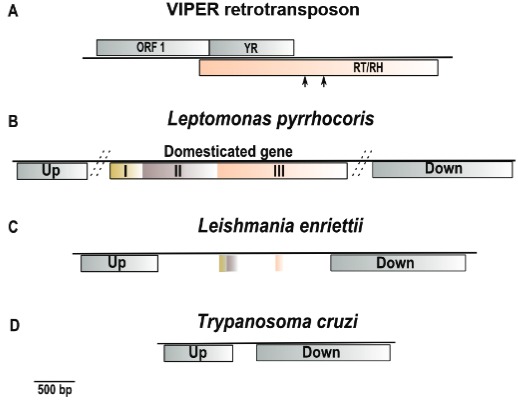
Fig. 1. : representation of the vestigial interposed retroelement (VIPER) retrotransposon, the domesticated gene, and the syntenic regions. (A) Schematic representation of the VIPER retrotransposon consensus sequence from Trypanosoma cruzi. It has 4390 bp, containing three genes, ORF1 from 169-1509, YR gene from 1510-2517 and RT/RH gene from 1386-4214. Arrows indicate alternative start sites for the third gene; (B) schematic representation of the Leishmania pyrrhocoris domesticated gene and its genomic context. The distinct gene regions I, II, and III are indicated with different box colours. The region III is homologous to the C-terminal portion of the third VIPER protein. Up and Down boxes on B, C, and D represent orthologous genes that are upstream and downstream the domesticated gene. The upstream gene has approximately 900 bp and is annotated for some species as “Zn-finger in Ran binding protein and others”. The downstream gene has approximately 1700 bp and is annotated as “hypothetical protein, conserved”. The TriTrypDB gene identification (ID) of these genes for each species can be found in Supplementary Table I (2.6MB, pdf) . Dotted bars in the intergenic regions represent scale discontinuity; (C) schematic representation of the L. entiettii syntenic region showing an intergenic region with remnants of the domesticated gene; (D) schematic representation of the T. cruzi syntenic region showing a short intergenic region without any evidence of the domesticated gene.