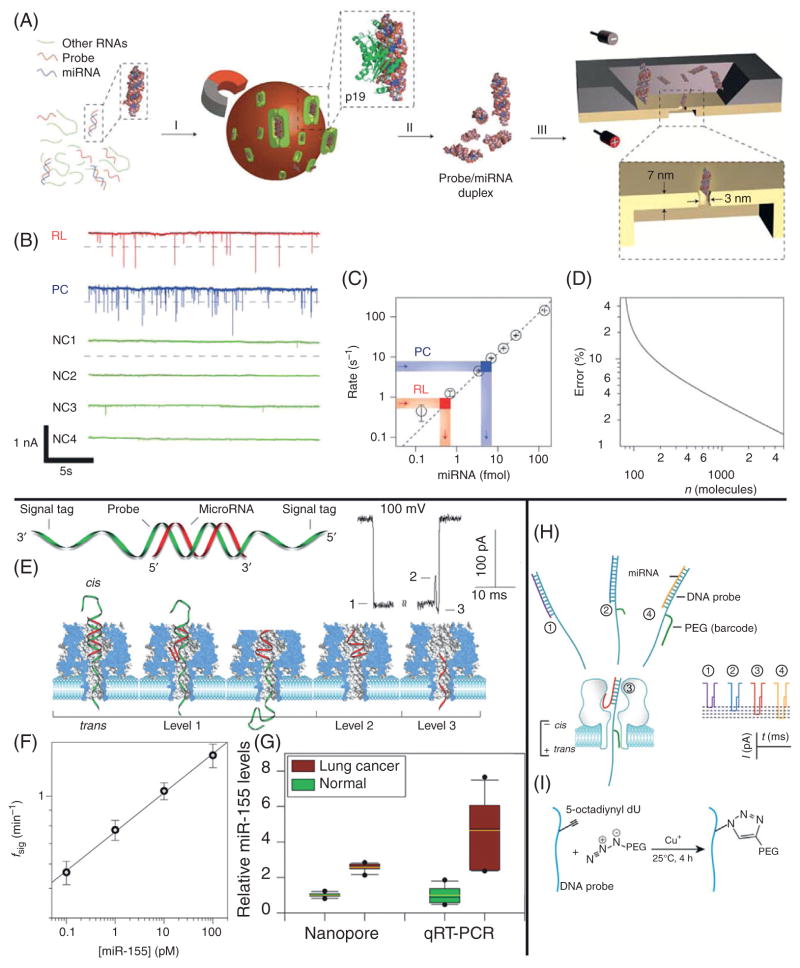
Figure 3.
Nanopore-based miRNA detection schemes. (A) A cartoon of the miRNA detection scheme used by the Drndić Lab.20 Total RNA extract is incubated with probe molecules complimentary to a target miRNA sequence. This mixture is then incubated with magnetic beads that are functionalized with the p19 protein, which selectively binds to 21–23 bp dsRNA molecules. A washing step then isolates the target miRNA/probe complex, which can be readily detected using a thin SiN nanopore. (B) A sample of the ionic current traces resulting from translocations of the isolated miRNA/probe complexes. RL is the trace resulting from enrichment of miR122a, PC is a positive control, and NC refers to various negative control experiments. (C) A calibration curve is constructed by plotting the event rates as a function of miRNA concentration. The event rates of the PC and RL experiments are shown as indicated and the miRNA concentration is inferred from the calibration curve. (D) The error in concentration as predicted from the nanopore experiments, as a function of number of translocation events. (E) A diagram of the unzipping method used by the Gu Lab.22 A probe/target miRNA complex is shown along with the translocation process of this molecule and its resulting ionic current trace. (F) A calibration curve constructed by plotting event rate versus miRNA concentration is shown. (G) The miRNA levels as detected by the nanopore, as well as qRT-PCR are compared. (H) A schematic of the revised Gu Lab strategy,23 employing four different polyethylene glycol (PEG) tags. (I) A diagram of the click chemistry tagging process. Images obtained with permission from Wanunu et al.20,22