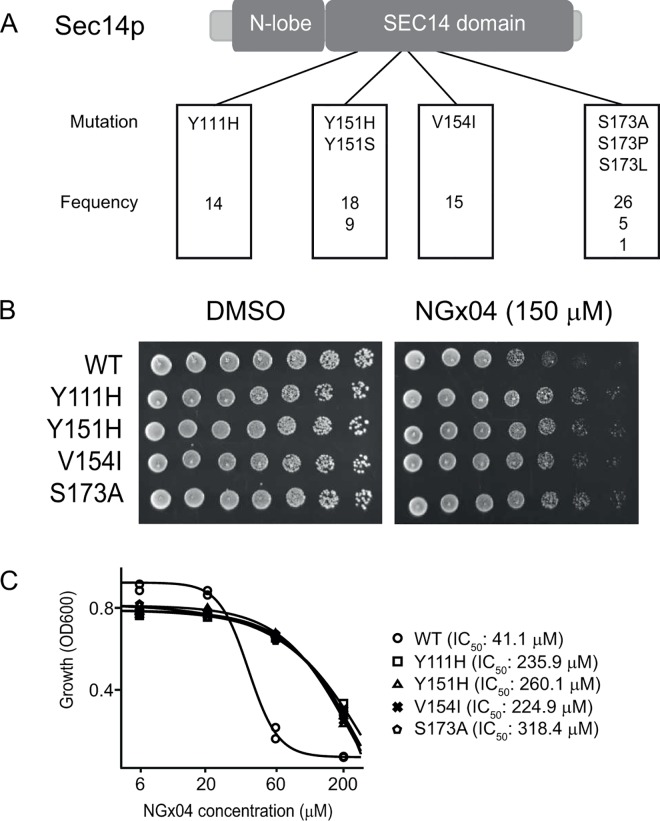
Fig 2. Resistance mapping.
Functional variomics screen for NGx04-resistant mutants. A) The SEC14 ORFs of 88 NGx04-resistant colonies were amplified by PCR and sequenced. Four mutations mapping to the lipid-binding part of Sec14p were found. The identified amino acid changes with their respective frequencies are given. B) The plasmids coding for various SEC14 proteins were re-transformed into a fresh wild type yeast strain and serial dilutions of the cultures spotted on either 150 μM of NGx04 or DMSO control, incubated for 3 days at 30°C and scored for growth. C) The strains were grown for 24 h in the presence of increasing concentrations of NGx04 and growth measured by turbidity (OD600). Duplicate values were determined and the 18 h time point was used for logistic regression. The calculated IC50 values displayed.