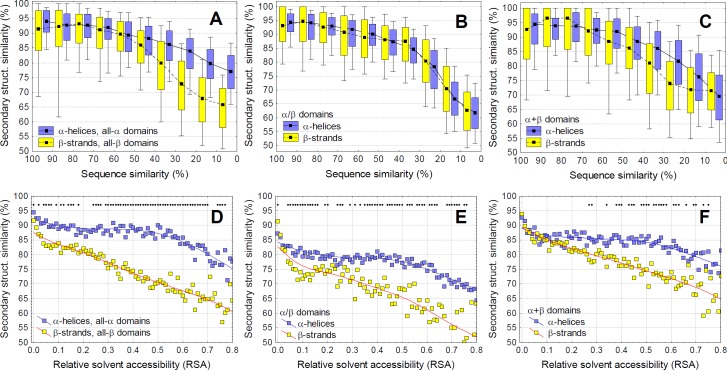
Fig 2. Alpha helices can accept more mutations than beta strands.
A-C) Secondary structure similarity of pairwise alignments as a function of sequence similarity. Pairwise alignments were grouped into 10% bins based on their sequence similarity. α-helices change significantly less with sequence change than beta strands in case of all-α, all-β and α+β domains. D-F) Since in α/β and α+β domains there is a large difference in the buriedness of helices and strands (see S1 Fig), using alignments with 10–20% similarity, we added relative solvent accessibility (RSA) as a covariate. When this correction is applied (i.e. the different levels of buriedness are taken into account), residues of helices are significantly more robust for mutations than strands in all SCOP classes, except for the most buried residues with RSA < 0.1 (diamonds indicate significant difference, tests of proportions, p < 0.05 after Holm-Bonferroni correction).