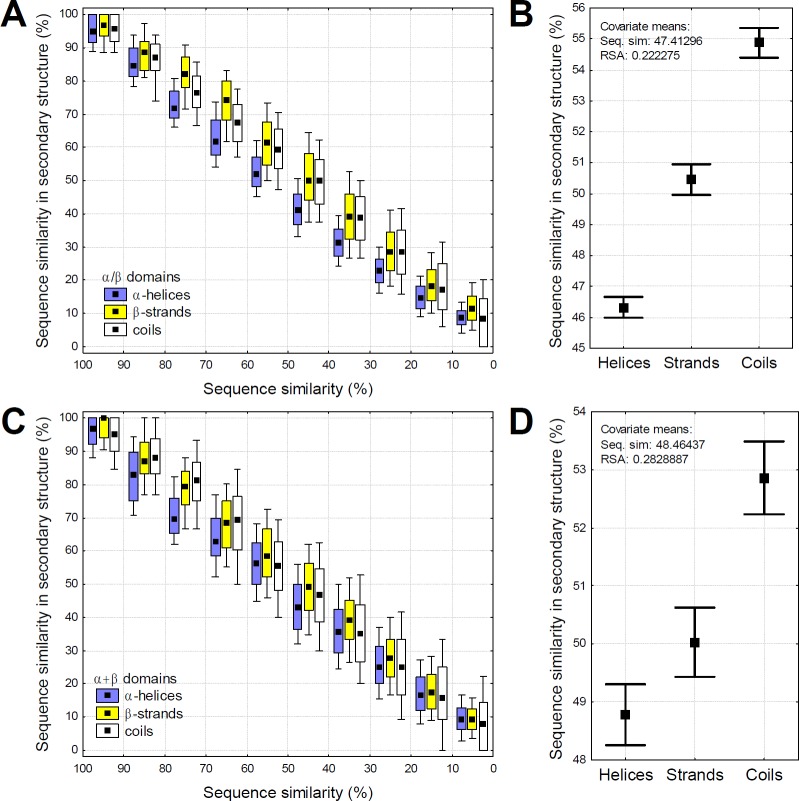
Fig 4. Within the same protein domains, helices diverge faster than strands, indicating higher robustness.
A) The relationship between global sequence similarity and sequence similarity in secondary structures in α/β domains. The pairwise structural alignments were grouped into 10% bins (see Fig 2), boxes represent 25–75%, whiskers 10–90%. Note that the difference between helices and strands declines below 40% sequence similarity, because sequence similarity cannot be negative, and random sequences have an expected similarity of 5–6%. In the alignments with 10–90% sequence similarity, helices are significantly more diverged than strands and coils in each bin (p< 0.05, t-tests), also when the differences in their RSA is taken into account (p< 0.05, ANCOVA). B) An example of the independent effect of secondary structure on sequence divergence in α/β domains, using the pairwise structural alignments with 40–60% divergence, and ANCOVA with global sequence similarity and the average RSA of secondary structure as continuous predictors. Within the same domains, helices are significantly more diverged than strands (p < 2 x 10−16, whiskers represent 95% confidence intervals), which in turn are more diverged than coils (p < 2 x 10−16). C-D) The same as A-B, but for α+β domains.