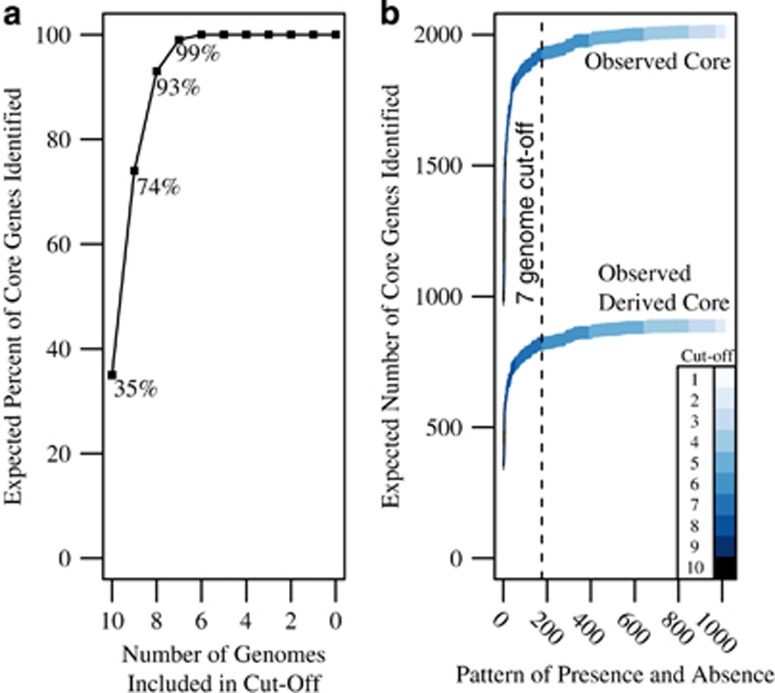
Figure 2.
(a) The expected percentage of core gene families identified for each pattern of presence and absence was calculated using the genome completeness estimates. Using these probabilities, a cutoff of seven genomes is expected to identify 99% of all core genes. This cutoff was used in conjunction with ancestral state reconstructions to determine the core genome of the Accumulibacter lineage. Only gene families that were inferred at the LCA of Accumulibacter and all internal nodes (for example, not lost until a terminal node) and were present in seven or more genomes were considered core in this analysis. (b) The observed number of core and derived core gene families using variable cutoffs. Each potential core gene family was sorted based on the number of genomes they were present in and then on the expected frequency of the pattern. Next, the cumulative sum of each additional pattern was calculated as patterns of increasing likelihood were added. The cutoff at seven genes is demarcated with a dotted line.