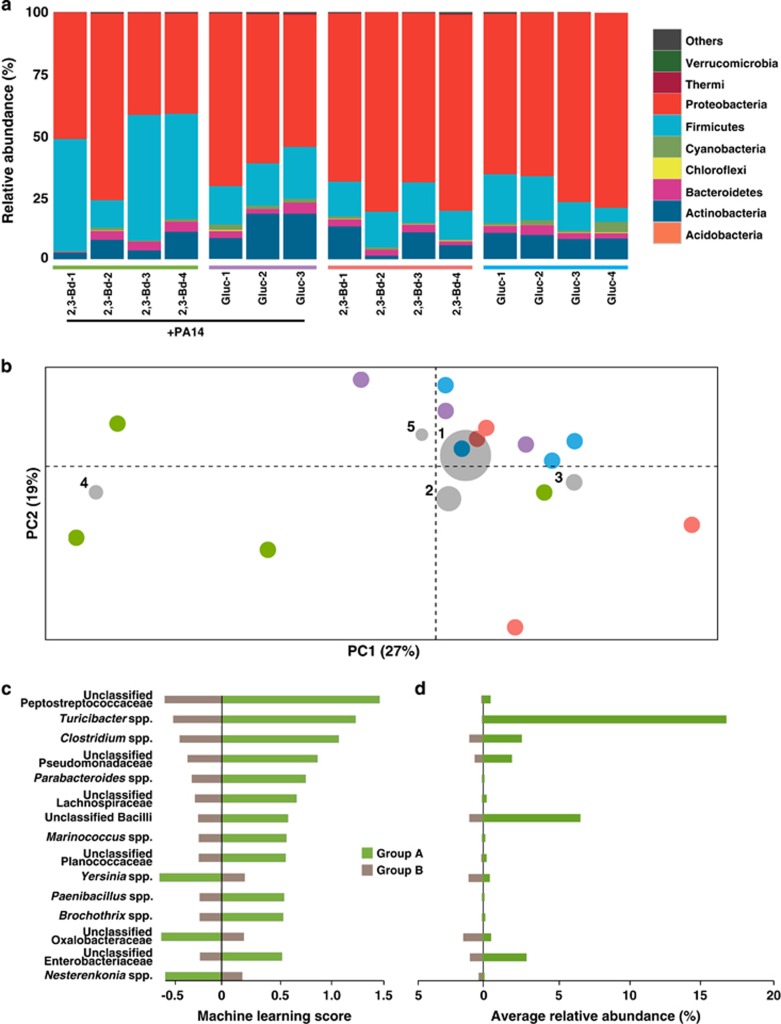
Figure 2.
Microbiome of the murine lung. (a) Taxonomic summary of lung microbiota. 16S rRNA gene sequences of lung bacterial community of 15 mice 3 days after infection with varying agar beads treatments, indicated by colored bars above the sample ID: PA14 with 2,3-butanediol (2,3-Bd+PA14, green); PA14 with glucose (Gluc+PA14, purple); 2,3-butanediol medium only (2,3-Bd, red); and glucose medium only (Gluc, blue). Each column represents a sample from one mouse. Bacteria are presented at the phyla level. (b) Principal Coordinates Analysis biplot of murine lung microbiomes. Beta diversity Principal Coordinates Analysis of a weighted UniFrac distance matrix from taxonomic composition within these samples. Only the first two axes are shown. Each circle represents one mouse, which is colored by agar beads treatment: PA14 with 2,3-butanediol (green; n=4); PA14 with glucose (purple; n=3); 2,3-butanediol medium only (red; n=4); and glucose medium only (blue; n=4). Gray circles represent taxa coordinates that are calculated based on mean relative abundance (size of circle) for each taxon in all 15 lung samples (1-Pseudomonas spp.; 2-Acinetobacter spp.; 3-Escherichia spp.; 4-Turicibacter spp.; 5-Staphylococcus spp.). The percentage of distribution described by each axis is as indicated. (c) Machine-learning analysis of murine lung microbiomes. Machine-learning analysis of lung samples binned into two groups based on agar treatment: Group A (2,3-butanediol with PA14; green; n=4) and Group B (glucose medium with PA14, 2,3-butanediol medium, and glucose medium; brown; n=11). Shown are the 15 highest correlated taxa (of the 593 taxa used to train the algorithm); the overall machine-learning error rate was 0.19 via nearest shrunken centroid method. (d). The average relative abundances of the 15 most predictive taxa from machine-learning analysis, and colored as in c.