Abstract
Microbial interactions, whether direct or indirect, profoundly affect the physiology of individual cells and ultimately have the potential to shape the biogeochemistry of the Earth. For example, the growth of Prochlorococcus, the numerically dominant cyanobacterium in the oceans, can be improved by the activity of co-occurring heterotrophs. This effect has been largely attributed to the role of heterotrophs in detoxifying reactive oxygen species that Prochlorococcus, which lacks catalase, cannot. Here, we explore this phenomenon further by examining how the entire transcriptome of Prochlorococcus NATL2A changes in the presence of a naturally co-occurring heterotroph, Alteromonas macleodii MIT1002, with which it was co-cultured for years, separated and then reunited. Significant changes in the Prochlorococcus transcriptome were evident within 6 h of initiating co-culture, with groups of transcripts changing in different temporal waves. Many transcriptional changes persisted throughout the 48 h experiment, suggesting that the presence of the heterotroph affected a stable shift in Prochlorococcus physiology. These initial transcriptome changes largely corresponded to reduced stress conditions for Prochlorococcus, as inferred from the depletion of transcripts encoding DNA repair enzymes and many members of the ‘high light inducible' family of stress–response proteins. Later, notable changes were seen in transcripts encoding components of the photosynthetic apparatus (particularly, an increase in PSI subunits and chlorophyll synthesis enzymes), ribosomal proteins and biosynthetic enzymes, suggesting that the introduction of the heterotroph may have induced increased production of reduced carbon compounds for export. Changes in secretion-related proteins and transporters also highlight the potential for metabolic exchange between the two strains.
Introduction
Most of our knowledge of microbial physiology is based on experiments with pure cultures, but these organisms did not evolve in isolation. Microbes can affect each other through a variety of positive (such as cooperation, symbiosis or mutualisms), negative (competition, antagonism) or neutral mechanisms that influence their function and behavior (Azam and Malfatti, 2007; Hibbing et al., 2010). Such interactions may occur either through direct physical contact or via diffusive signals across large spatial or temporal distances (Malfatti and Azam, 2009). Although many types of interactions are known to occur, the ways in which such interactions specifically alter cellular physiology remain poorly characterized.
Microscale interactions among marine microbes have the potential to impact both cellular physiology and, ultimately, influence global biogeochemical cycles (Azam and Malfatti, 2007). Interactions between autotrophs and heterotrophs are central to the marine food web, as heterotrophic bacteria are dependent on photosynthetically fixed organic carbon supplied by primary producers. Autotroph–heterotroph interactions have been shown to impact the autotroph as well, as work in the cyanobacterium Synechococcus has shown that co-culture impacts many cellular processes including carbon metabolism and stress responses (Tai et al., 2009; Beliaev et al., 2014). Growth of Prochlorococcus—the numerically dominant phytoplankter in the world's oceans (Flombaum et al., 2013)—in culture is improved by the presence of certain heterotrophs in terms of longevity, stationary phase cell density and ability to grow from low cell density (Sher et al., 2011). This phenomenon is attributable at least in part to the role of heterotrophs in reducing the levels of toxic reactive oxygen species such as hydrogen peroxide (Morris et al., 2008, 2011). This is thought to compensate for the absence of catalase in the Prochlorococcus genome, indicating that interspecies interactions have clearly influenced the evolutionary selective pressures and processes acting upon this group of organisms (Morris et al., 2012). Heterotrophs in Prochlorococcus cultures in turn receive photosynthetically fixed organic carbon and other nutrients supplied by Prochlorococcus; indeed, many heterotrophs thrive in Prochlorococcus co-cultures in the absence of any added carbon. Prochlorococcus cultures have been historically difficult to render axenic, likely due in part to this intimate co-dependency (Chisholm et al., 1992; Moore et al., 2007). Not all Prochlorococcus—heterotroph interactions are mutually beneficial, however. Some heterotroph strains can either inhibit Prochlorococcus growth or have no obvious impact in co-culture conditions, suggesting the existence of both general and strain-specific interaction outcomes (Sher et al., 2011).
The physiological mechanisms underlying the different types of interrelationships that occur among Prochlorococcus and other microbes undoubtedly extend far beyond reactive oxygen species scavenging and fixed carbon exchange (Becker et al., 2014). As a step toward further understanding the underpinnings of these interactions, we examined the transcriptional responses of Prochlorococcus strain NATL2A to growth in pure culture vs in co-culture with the marine heterotroph, Alteromonas macleodii strain MIT1002 (Biller et al., 2015b) (hereafter ‘Alteromonas'). These strains were previously sympatric, having been co-isolated from the same water sample and co-cultured together for many years (along with other naturally co-occurring heterotrophs). By reuniting these two strains and looking at their initial transcriptomic reactions, we sought to gain insights into the types of interactions that may have facilitated their sustained co-existence. How long does it take for Prochlorococcus to ‘notice' the presence of heterotrophs after they are introduced? What gene expression changes result, and what cellular processes may have been impacted? Are these transient responses, or do they suggest that co-culture conditions may induce a stable shift in cellular physiology?
Materials and methods
Culturing and sampling
Prochlorococcus NATL2A was originally isolated from 10 m depth in the North Atlantic (Scanlan et al., 1996) and is a member of the low-light I clade of Prochlorococcus (Biller et al., 2015a). Axenic Prochlorococcus NATL2A cells were grown in natural sea water based Pro99 media containing 0.2 μm filtered Sargasso Sea water amended with Pro99 nutrients prepared as previously described (Moore et al., 2007), except with the addition of 10mm sterile sodium bicarbonate to support the growth of a large volume of culture (Moore et al., 2007). Prochlorococcus cultures were grown at 21 °C under constant illumination (30 μmol photons m−2 s−1). A. macleodii MIT1002 (Biller et al., 2015b) was maintained at 21 °C in ProMM medium (Pro99 media, as above, plus lactate, pyruvate, glycerol, acetate and Va vitamins) (Berube et al., 2015).
To initiate the co-culture, triplicate 900 ml starter cultures of axenic Prochlorococcus NATL2A were grown in Pro99 as above, then split to form six replicate 450 ml cultures. When cultures reached mid-exponential growth phase, exponentially growing Alteromonas cells were spun down, washed twice with Pro99 media (to minimize any carryover) and added at a ~1:1 Prochlorococcus:heterotroph cell ratio (as determined by flow cytometery) to three of the co-culture bottles. An equivalent volume of sterile Pro99 media was added to the other three cultures that remained axenic. Bulk chlorophyll fluorescence and flow cytometry samples were collected daily to monitor cell abundance; Prochlorococcus were counted based on chlorophyll fluorescence, and heterotrophs were enumerated following SYBR staining. The purity of axenic NATL2A cultures was verified before and after the experiment by testing for growth in three purity broths (ProAC, ProMM and MPTB (Saito et al., 2002; Morris et al., 2008; Berube et al., 2015)) and by flow cytometry. Transcriptome samples were collected by removing 10 ml of culture and placing it immediately into 30 ml of cold RNALater, then incubating these samples for 1–3 days at 4 °C. Following RNALater incubation, cells were filtered onto 25 mm 0.2 μm Supor filters (Pall) and frozen at −80 °C.
RNAseq library construction
To extract total RNA, filters containing the cell biomass were first incubated in 10 mm Tris pH 8, 20 KU of Ready-Lyse lysozyme (Epicentre, Madison, WI, USA) and 40 U of SUPERase-In RNase inhibitor for 5min at room temperature. RNA was then extracted using the mirVana miRNA extraction kit (Ambion, Carlsbad, CA, USA) according to the manufacturer's instructions. Strand-specific RNAseq libraries were constructed with the dUTP second-strand approach as in (Giannoukos et al., 2012), and the ribosomal RNA content of each library was reduced using duplex-specific normalization (Yi et al., 2011; Giannoukos et al., 2012). Detailed sequencing library construction protocols can be found in Supplementary Methods. Libraries were sequenced on an Illumina HiSeq2000 at the MIT BioMicro Center, yielding ~1.9–3.5 million 40+40nt paired-end reads per library (Supplementary Table S1). The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Barrett et al., 2013) and are accessible through GEO Series accession number GSE73511 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73511).
Sequence analysis
Low-quality sequence regions were removed from the raw Illumina data using quality_trim (from the CLC Assembly Cell package, CLC bio) with default settings (-c 20 –b 0.1 –l 0.5). The paired-end RNAseq reads were aligned with the Burrows-Wheeler Aligner (Li and Durbin, 2009) to a reference file containing both the Prochlorococcus NATL2A and A. macleodii MIT1002 genomes (NCBI GenBank accessions CP000095 and JXRW01000000, respectively). The resultant alignments were parsed with aid of the samtools package (Li et al., 2009) using custom Python scripts to determine the number of paired-end fragments aligned to each annotated ORF in the sense orientation. The NATL2A gene calls and annotations used were made by the RAST pipeline (Aziz et al., 2008) and previously described (Biller et al., 2014); the A. macleodii MIT1002 genome was annotated in the same way for consistency (data available from GEO). In some cases, RAST predicted NATL2A coding sequences not present in the GenBank entry for the genome, and these are listed solely by chromosomal location (Supplementary Table S2).
Identification of significantly differentially abundant transcripts was performed with the DESeq2 R package (V1.6.3) (Love et al., 2014), using standard DESeq2 functions and workflows as recommended by the authors. Raw counts of reads mapping to each ORF (omitting RNAs) across all time points and replicates were first compiled; reads mapping to the Prochlorococcus and A. macleodii MIT1002 genomes were stored in separate tables and analyzed independently. Next, we estimated size factors for each sample to normalize for differences in library sequencing depth. Results from the biological replicates were used to estimate the dispersion of each gene. Tests for differential expression were carried out between each pairwise condition of interest (that is, the co-cultured and axenic Prochlorococcus samples from a given time point) with the Wald test, using a negative binomial generalized linear model. Resulting P-values were adjusted for multiple testing using the Benjamini—Hochberg procedure. Transcripts with an adjusted P-value<0.1 were considered to be differentially abundant (Love et al., 2014).
Analysis of pathway-level changes in gene expression was carried out with PathwayTools V18.5 (Karp et al., 2010; Caspi et al., 2014), using either the Prochlorococcus NATL2A Pathway/Genome Database or one generated for A. macleodii MIT1002. Relevant lists of genes previously found by DESeq2 to be differentially abundant (that is, all genes found to be differentially abundant in Prochlorococcus by DESeq2 at any time point in co-culture) were imported as SmartTables in PathwayTools. Pathway enrichment/depletion was tested using default settings (Fisher's exact test P-value <0.1 (Rivals et al., 2007)).
Results and Discussion
Impacts of co-culture on growth dynamics
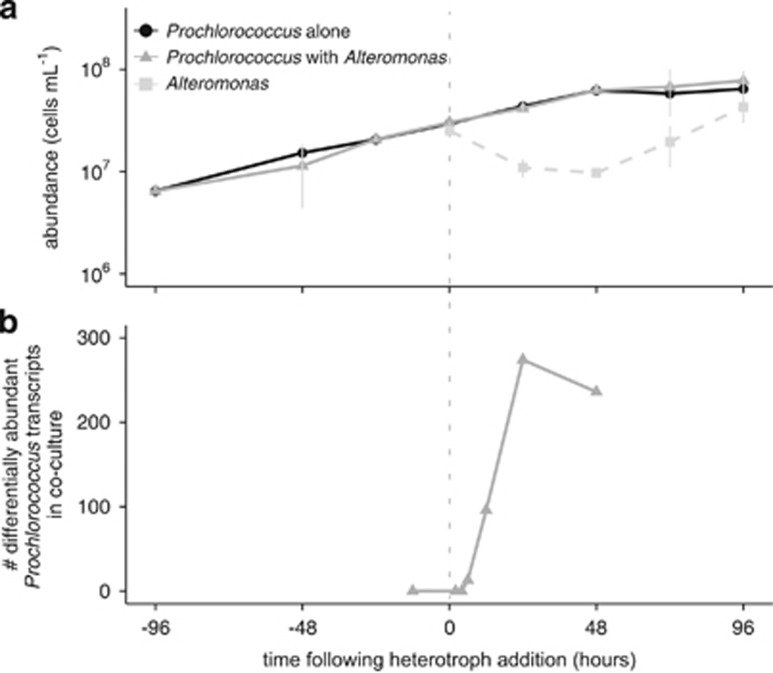
In order to mimic natural marine conditions, where heterotroph growth depends on the photosynthate released by organisms like Prochlorococcus, the co-culture experiment was conducted in Pro99 growth media, which lacks any added organic carbon sources or vitamins (Moore et al., 2007). The addition of the heterotroph (at a ~1:1 cell ratio) to an exponentially growing Prochlorococcus culture had no significant impact on Prochlorococcus growth rates (~0.37 per day) or final stationary cell density (Figure 1a). Prochlorococcus cells continued to grow exponentially for 2 days following heterotroph addition, after which they began to enter stationary phase. Flow cytometry analysis showed that Prochlorococcus cell size (as estimated from forward scatter measurements) and average fluorescence per cell did not change following heterotroph addition (Supplementary Figure S1). Heterotroph cell abundances declined for the first day following their introduction to the culture, were stable the following day, and then began to grow (Figure 1a). It is unclear why this initial decline occurred; the cell loss could have resulted in part from physical stress suffered upon washing the cells prior to adding them to the co-cultures, or may reflect nutrient limitation(s) that were unable to support that population size. The lag in growth might reflect either the time it took the Alteromonas to adjust to the utilization of organic carbon supplied by Prochlorococcus, as their previous sources of organic carbon were removed during the transfer to the NATL2A culture, or perhaps suggest a delay in the availability of those nutrients. The fact that the presence of this Alteromonas does not affect NATL2A exponential growth rates under near-optimal light and temperature conditions (Figure 1a) ensured that any transcriptional responses observed in NATL2A under co-culture conditions could be more directly attributed to the presence of the heterotroph and not complicated by changes in cellular growth rate.
Figure 1.
Impact of the introduction of a heterotroph on Prochlorococcus cell abundance and gene expression. (a) Cell abundances of axenic (black line, circles) and co-cultured (solid gray line, triangles) Prochlorococcus NATL2A, as well as Alteromonas macleodii MIT1002 in the co-cultures (dashed grey line, squares) during the experiment. Alteromonas cells were added to the culture at the ‘0' hour time point (vertical dashed line). Values represent the mean (±s.d.) of three biological replicates. (b) The number of differentially abundant transcripts in co-cultured vs axenic Prochlorococcus. Co-cultured and axenic samples were compared pairwise for each indicated time point.
Timing and general patterns of the Prochlorococcus transcriptional response to co-culture
To gain insight into how Prochlorococcus responded physiologically to the abrupt appearance of the heterotroph, we compared the transcriptomes of Prochlorococcus/Alteromonas co-cultures with Prochlorococcus growing alone (Figure 1). Samples were collected following 2, 4, 6, 12, 24 and 48 h of co-culture to examine both the initial and longer-term changes in transcriptional patterns (Table 1). The transcriptome of Prochlorococcus began to change between 4 and 6 h after the initial exposure to the heterotroph (Figure 1b, Table 1), which represents ~10% of the Prochlorococcus doubling time (~1.8 days) in this experiment. The number of differentially abundant transcripts continued to increase over the next day, reaching a peak of 274 genes (~12% of the NATL2A genome) 24 h after the heterotroph addition (Figure 1b). In total, transcripts representing 375 different Prochlorococcus genes were found to be differentially abundant during at least one time point in the co-culture, with more decreasing than increasing in relative abundance. The former was particularly evident at the early time points; for example, all of the differentially abundant transcripts after 6 h, and 88/96 (92%) of those after 12 h, were less abundant in co-cultured cells than in the axenic cultures (Table 1).
Table 1. Details of changes in the Prochlorococcus transcriptome during co-culture.
| Time after heterotroph addition (h) | Increased relative abundance | Decreased relative abundance | ‘Core' Prochlorococcus genes | Remained differentially abundant throughout the following time points |
|---|---|---|---|---|
| 2 | 0 | 0 | ||
| 4 | 0 | 0 | ||
| 6 | 0 | 13 | 12 | 13 |
| 12 | 8 | 88 | 45 | 63 |
| 24 | 120 | 154 | 149 | 162 |
| 48 | 101 | 135 | 108 |
Values indicate the number of genes with significantly differentially abundant transcript levels in the Prochlorococcus+Alteromonas co-culture transcriptome as compared with axenic Prochlorococcus at each time point.
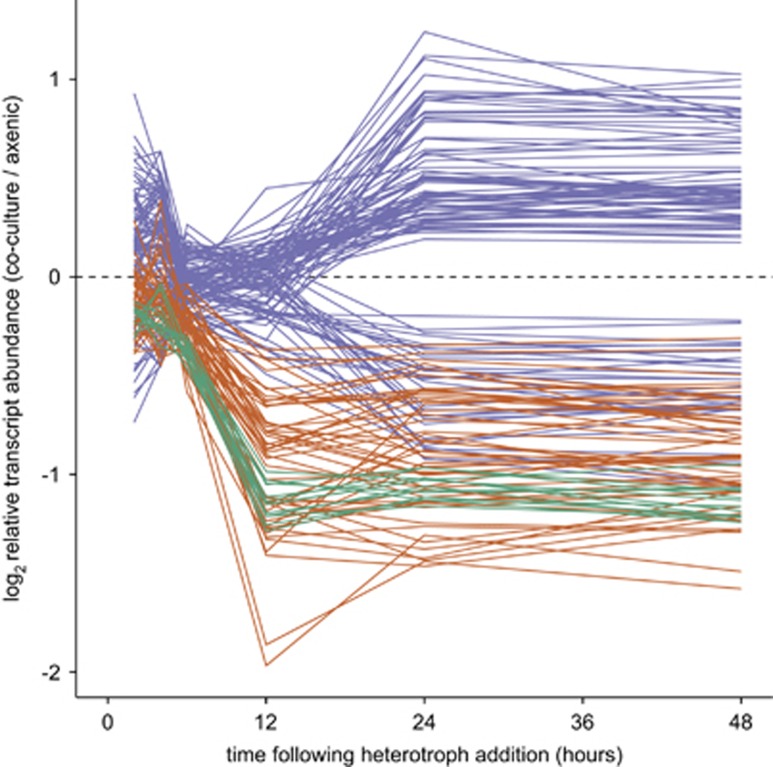
Changes within the Prochlorococcus transcriptome in response to the addition of the heterotroph occurred in stages, and many of the differences persisted throughout the experiment (Figure 2). The earliest response consisted of 13 transcripts that decreased in relative abundance beginning 6 h after heterotroph addition; all of them remained significantly differentially abundant throughout the rest of the time course (green lines, Figure 2). In the second stage, beginning after 12 h of co-culture, an additional 50 transcripts decreased in relative abundance and remained so for the duration of the experiment (red lines, Figure 2). After 24 h of co-culture, a further 99 transcripts (purple lines, Figure 2) were first observed to be significantly differentially abundant, and then remained consistently changed. Thus the addition of Alteromonas to the axenic Prochlorococcus culture resulted in a series of transcriptional changes that occurred in distinct temporal waves, with ~43% of the transcripts whose abundance changed in co-culture at any time point did so in a non-transient manner. This pattern suggests that co-culture conditions resulted in persistent physiological changes within Prochlorococcus.
Figure 2.
Persistent changes in the Prochlorococcus NATL2A transcriptome in the presence of a heterotroph. Each line represents the relative abundance of a transcript that remained significantly differentially abundant in co-cultured cells throughout the duration of the experiment. Colors indicate groups of transcripts that first became significantly differentially abundant, 6 (green), 12 (red) or 24 (purple) hours after addition of Alteromonas macleodii MIT1002.
More than half (192) of the 375 differentially abundant transcripts represent ‘core' genes in Prochlorococcus (Table 1); that is, genes with homologs in all 41 sequenced Prochlorococcus genomes to date (Biller et al., 2014). However, a number of the differentially abundant transcripts—and, indeed, many of those showing the greatest magnitude change—encode proteins with no currently annotated function (Supplementary Table S2). This is perhaps not surprising given the fact that ~75% of all known Prochlorococcus genes are unannotated (Biller et al., 2015a), but the lack of annotation hampers the interpretation of many of the responses observed.
Co-culture appears to reduce cellular stress conditions in Prochlorococcus
Many of the transcriptional responses of Prochlorococcus can be associated with reduction of cellular stress under co-culture conditions. Of the first 13 transcripts that changed in abundance 6 h after heterotroph addition, 12 encoded members of the high light inducible (hli) family of stress-responsive proteins (the other is of unknown function; Supplementary Table S2). hlis were originally identified as genes that played a role in protecting cells from light shock (Funk and Vermaas, 1999; He et al., 2001; Bhaya et al., 2002), and are now known to respond to a variety of stresses in Prochlorococcus (Tolonen et al., 2006; Lindell et al., 2007; Thompson et al., 2011; Bagby and Chisholm, 2015). hlis are found in all Prochlorococcus genomes, but NATL2A is notable for having a much larger suite of hlis than many other Prochlorococcus (Coleman and Chisholm, 2007; Kettler et al., 2007; Berta-Thompson, 2015), which may be related to the ability of this clade of low-light adapted Prochlorococcus to withstand transient exposure to high light intensities (Malmstrom et al., 2010). All 12 of these hli transcripts decreased in relative abundance in co-culture conditions, suggesting that the Alteromonas may have mitigated some form of stress in the culture, such as oxidative stress (Morris et al., 2011). These ‘early response' hli transcripts were also found clustered within four sets of adjacent operon-like hli arrays in the genome (arrays 1, 4, 5 and 6; Supplementary Table S2), suggesting that these transcripts may be co-regulated. Transcripts encoding an additional 19 hlis decreased in abundance after 12 h of co-culture, and three more decreased after 24 h. After the 24 h time point there was one hli transcript—PMN2A_0886—that increased in relative abundance in the presence of Alteromonas. In total, after the initiation of co-culture the relative transcript levels of 36/42 (86%) of all hli genes in the NATL2A genome changed during at least one time point.
Additional transcriptional responses of Prochlorococcus were consistent with a generalized reduction in oxidative stress in the presence of the heterotroph (Morris et al., 2011). For example, multiple DNA repair enzymes, including the DnaJ chaperone, a putative deoxyribodipyrimidine photolyase, the RecG helicase and the error-prone DNA repair protein UmuD were transcriptionally depleted in co-culture (Table 2). In addition, other stress-associated transcripts encoding the ClpB chaperone (which helps to refold stress-damaged proteins), transcripts for purine nucleotide salvage, an alkyl hydroperoxide reductase and multiple putative oxidoreductases also decreased in relative abundance (Supplementary Table S2). The degree of oxidative stress experienced by the axenic culture was apparently insufficient, however, to impact growth rates (Figure 1a) or the expression of transcripts encoding the PSII D1 and D2 proteins, which are sensitive to oxidative damage (Mella-Flores et al., 2012).
Table 2. Prochlorococcus pathways and selected GO categories significantly enriched or depleted in co-culture.
| Pathway | Genes with significantly differentially abundant transcript levels | P-value |
|---|---|---|
| Increased abundance during co-culture | ||
| Vitamin B12 biosynthesis | PMN2A_0380, PMN2A_1611, PMN2A_1855 | 7.4E-02 |
| Transcription regulation | nrdR, nusG, PMN2A_0013, PMN2A_1305, PMN2A_1829 | 1.3E-03 |
| Generation of precursor metabolites and energy | chlL, pgi, PMN2A_1150, PMN2A_1308, PMN2A_1659, PMN2A_1660, PMN2A_1801, PMN2A_1851 | 1.4E-02 |
| Cofactor biosynthesis (including chlorophyll) | chlL, chlN, nadC, tal, PMN2A_0164, PMN2A_0380, PMN2A_0840, PMN2A_1583, PMN2A_1865 | 1.6E-02 |
| Photosynthesis | chlL, chlN, petA, psaD, psaF, psaI, psaJ, psaL, psbF, ycf3, PMN2A_0164, PMN2A_1678 | 3.6E-05 |
| NAD Metabolism/biosynthesis | nadC, nadD | 7.7E-02 |
| Intracellular transport | secA, secE | 9.3E-02 |
| Decreased abundance during co-culture | ||
| Amines and polyamines degradation; urea degradation | ureA, ureB, PMN2A_1862 | 5.7E-03 |
| Nucleic acid metabolism | dnaB, dnaJ, gidB, lysS, phr, recG, rlmH, rne, umuD, PMN2A_0696, PMN2A_0726, PMN2A_0812, PMN2A_1207, PMN2A_1216, PMN2A_1292, PMN2A_1537 | 2.9E-02 |
| Nucleosides and nucleotides degradation | PMN2A_0143, PMN2A_1666 | 8.1E-02 |
| Purine riboucleotides biosynthesis | purH, PMN2A_0117 | 4.2E-02 |
This analysis is based on the set of transcripts that were found to be significantly differentially abundant in the transcriptome dataset in at least one time point following heterotroph addition (see Supplementary Table S2). P-value indicates the result of a Fisher's significance test (PathwayTools).
Transcriptional changes in photosynthetic machinery and electron transport
While many of the earliest transcriptional responses to co-culture involved decreases in transcript abundance, after 24 h a number of Prochlorococcus transcripts began to increase in relative abundance. A number of these encoded proteins related to photosynthesis, including photosystem components, cytochromes and cytochrome oxidase, and a phycoerythrin subunit (PMN2A_1678) (Table 2). Transcripts for chlorophyll biosynthesis enzymes also increased, but curiously there was no significant change in red fluorescence per cell (Supplementary Figure S1). Although the latter is often used as a proxy for cellular chlorophyll content, chlorophyll fluorescence is a function of many variables and this lack of relationship may not necessarily be surprising. We observed that transcripts for numerous PSI subunits, as well as the PSI assembly protein Ycf3, were enriched during co-culture, while only two PSII-related proteins (the cytochrome b559 subunit PsbF and a PSII assembly protein, Psb27) showed a significant response (Table 2). PSI:PSII ratios have been found to vary during the exponential growth of Prochlorococcus cultures (Zorz et al., 2015), but these changes were identified as significant across three biological replicate axenic and co-cultures. Further, given the similarity in growth rates and flow cytometry parameters among the axenic and co-cultured cells (Figure 1a, Supplementary Figure S1), these data suggest that this reshaping of the photosynthetic apparatus was a consequence of co-culture. The transcriptome data provide conflicting signs as to whether or not the ‘dark reactions' of photosynthesis involving carbon fixation also increased during co-culture. For instance, although transcripts encoding multiple carboxysome shell proteins were enriched, the transcriptional regulator for the RuBisCO operon (rbcR, PMN2A_1513) simultaneously decreased in abundance (though without any significant changes in transcript abundance for the RuBisCO subunits; Supplementary Table S2).
Why might the presence of a heterotroph trigger a change in the photosystems of Prochlorococcus? One possible explanation is that the heterotroph, by consuming some form of dissolved organic carbon released by Prochlorococcus, might have either directly or indirectly stimulated Prochlorococcus to increase its production of reduced carbon for export. In this case, by increasing PSI abundances, Prochlorococcus could generate additional ATP via cyclic electron flow around PSI to be used for carbon fixation or anabolic reactions. It is interesting to note that the growth of Alteromonas began when the Prochlorococcus population began to enter stationary phase (Figure 1a), when more of the captured light energy could perhaps have been directed toward overflow metabolism and used to fix carbon that was ultimately excreted into the media (Bertilsson et al., 2005). An alternative interpretation would be that the heterotroph shaded the Prochlorococcus cells, reducing the available light energy and resulting in changes in photosystem architecture in an attempt to increase light harvesting. We think the latter is unlikely because if the Prochlorococcus cells were experiencing significantly different light intensities in the presence of the heterotroph, we would expect to see changes in the flow cytometry fluorescence signatures (Supplementary Figure S1), as well as in the abundance of more PSII subunit transcripts—which we did not.
Changes in the photosystem could also reflect a cellular response to shifts in the redox state of the quinone pool, perhaps linked to the apparent reduction in cellular oxidative stress. A number of transcriptional responses to co-culture were suggestive of this type of change. For example, we observed significant enrichment for transcripts encoding proteins involved in photosynthetic electron transfer, such as for subunits of NAD(P)H-quinone oxidoreductase and NADH-ubiquinone/plastoquinone (Supplementary Table S2). Transcripts encoding the plastiquinol terminal oxidase (PTOX) were also less abundant following 24 and 48 h of co-culture. PTOX transfers electrons from plastoquinone to molecular oxygen to form water, and is thus involved in photoacclimation and maintaining cellular redox poise during high light conditions. Indeed, decreases in Prochlorococcus PTOX expression have been observed in 3-(3,4-dichlorophenyl)-1,1-N-N'-dimethylurea (DCMU)-treated cells (Steglich et al., 2006); DCMU is an inhibitor of PSII-mediated electron transport that causes a net oxidation of the plastoquinone pool. In addition, Prochlorococcus PTOX expression has been shown to increase during conditions of CO2 limitation (Bagby and Chisholm, 2015), reinforcing the role of this enzyme as a ‘safety valve' for the cell and implicating the decrease in PTOX transcript abundance seen here in co-culture as a reflection of generally reduced stress on the cells. PTOX and PSI components exhibit inverse expression patterns in Prochlorococcus experiencing iron starvation, with PTOX expression increasing while PSI is downregulated (Thompson et al., 2011). As the culture media in our experiments contained ample iron and there was no reason to think that co-culture changed iron availability, this observation reinforces the inverse relationship between these two systems in modulating photosynthetic electron flow.
Although we did not observe a change in the growth rate of axenic and co-cultured Prochlorococcus (Figure 1a), we did find that co-cultured cells were enriched for transcripts encoding a number of proteins involved in biosynthesis and/or growth. These included increases in transcripts for multiple ribosomal proteins, as well as enzymes involved in amino acid biosynthesis pathways (specifically genes in the chorismate pathway, which makes the precursor for tryptophan, tyrosine and phenylalanine biosynthesis). In addition, transcripts encoding multiple tricarboxylic acid cycle enzymes, glycolysis enzymes (including both a glycogen debranching enzyme and glucose-6-phosphate isomerase) and NAD biosynthesis enzymes were also enriched in the presence of the heterotroph. DnaE, a subunit of DNA polymerase III, exhibited increased transcript abundance across multiple time points as well (although the DnaB replicative helicase was transcriptionally depleted) (Supplementary Table S2). This suggests that Prochlorococcus cells were increasing their biosynthetic capability in co-culture, perhaps required to support the increased synthesis of proteins from the enriched transcripts.
Potential for metabolic exchange between Prochlorococcus and Alteromonas
Transfer of fixed organic carbon from the autotroph to the heterotroph is expected in autotroph–heterotroph co-cultures, but many other types of biomolecules might be exchanged as well. Indeed, transcriptional analysis of a Synechococcus–Shewanella co-culture found that 14% of the changes in the Synechococcus transcriptome were affiliated with transport processes (Beliaev et al., 2014). Similarly, after 1 day of co-culture the relative transcript abundance of components for at two ABC-type transporters (PMN2A_1780 and PMN2A_1891) increased in Prochlorococcus, whereas another, PMN2A_1455, decreased. A putative Mn2+/Fe2+ transporter, PMN2A_0237, was also significantly transcriptionally enriched 12 h after heterotroph addition, possibly related to the increased need for iron in cytochromes and PSI. In addition, transcripts encoding the secreted preprotein translocase component SecA, the SecE component of the preprotein transport channel, and a putative outer membrane transporter (PMN2A_1465) increased, suggesting the potential for increases in extracellular protein export during co-culture. Transcripts for a putative outer membrane import channel (PMN2A_0909) also significantly increased during co-culture (Supplementary Table S2). These responses suggest that the ability of Prochlorococcus to transport components either to or from the extracellular environment was enhanced following the addition of Alteromonas to the medium—whether due to a specific ‘recognition' of the heterotroph, or perhaps reflecting increased competition for nutrients.
Prochlorococcus NATL2A contains the complete pathway for synthesizing vitamin B12 (Kettler et al., 2007). Many marine microbes are known to be vitamin B12 auxotrophs as they require this cofactor for essential functions such as methionine biosynthesis, yet lack the capability to make it themselves (Croft et al., 2005; Sañudo-Wilhelmy et al., 2013); this has led to the suggestion that exchange of vitamin B12 may have an important ecological role in the marine environment (Kazamia et al., 2012; Bertrand et al., 2015). Owing to its massive population size, Prochlorococcus may be a globally important supplier of this cofactor for the ocean ecosystem (Bonnet et al., 2010). In our work we found that co-cultured Prochlorococcus exhibited increased abundance of transcripts encoding multiple B12 synthesis proteins as compared with axenic controls (Table 2; Supplementary Table S2). The genome of A. macleodii MIT1002 (Biller et al., 2015b) indicates that this strain does not need vitamin B12 for growth, since it encodes the B12-independent MetE methionine synthetase. However, MIT1002 has the potential capability to utilize vitamin B12 facultatively—even though it cannot produce this compound itself—as it also encodes the B12-dependent MetH methionine synthetase, an isoform of MetE. Although we do not know which form was being used by Alteromonas in co-culture, MetH is typically used preferentially in the presence of vitamin B12 (Helliwell et al., 2011; Bertrand and Allen, 2012), presumably due to the >100-fold higher catalytic efficiency of MetH over MetE (Gonzalez et al., 1992). Thus Prochlorococcus may have directly or indirectly provided Alteromonas with vitamin B12 in co-culture to facilitate growth of the ‘helper', although these transcriptional changes could also represent increased production for its own use due to other changes in its metabolism.
Curiously, our data also suggest that the presence of the heterotroph impacted phosphorous metabolism in Prochlorococcus. Transcripts for the PhoB transcription factor, which regulates expression of phosphate acquisition genes, were significantly less abundant in Prochlorococcus cells starting after 12 h of co-culture, when the cells were still in exponential phase and excess phosphorus was available (Figure 1a); phoB transcripts remained transcriptionally depleted throughout the rest of the experiment. Similarly, the phosphate starvation-inducible protein PhoH was also significantly less abundant after both 24 and 48 h of co-culture—the former when the cells were still in exponential growth phase and the latter at beginning of stationary phase. This suggests that phosphorous became more readily available to Prochlorococcus in the co-cultures well before they moved toward stationary phase and stayed that way. One possibility is that the Alteromonas cells might have released some additional P into the media through lysis, as their cell numbers decreased for about 24 h upon introduction to the co-culture; they began to grow exponentially again after 48 h (Figure 1a). Alternatively, the Alteromonas might have directly secreted phosphorous compounds from some form of internal stores into the culture. Perhaps relatedly, two urease subunits (ureA, ureB) were also transcriptionally depleted after 48 h in co-culture (Supplementary Table S2), suggesting changes to N metabolism or the recycling of N-containing compounds within Prochlorococcus as the co-culture began to approach stationary phase.
These observations collectively indicate that metabolic exchange between Prochlorococcus and this strain of Alteromonas may have occurred in both directions. Prochlorococcus supplied the heterotroph with fixed carbon and perhaps other essential compounds, and the presence of this sympatric Alteromonas may have made essential nutrients more available to Prochlorococcus through some mechanism. Although Prochlorococcus was eventually limited by some media component as it transitioned into stationary phase, the limiting nutrient(s) for each strain—and the degree to which they may have been competing for resources—are not known. Some of the changes in transporter transcripts may have ultimately helped Prochlorococcus to sequester additional nutrients in the face of increased competition with Alteromonas. By comparison, a previous study of Synechococcus WH8102 co-cultured with Vibrio noted an increase in expression of phosphate acquisition genes within Synechococcus even though phosphate was not depleted (Tai et al., 2009). That response was attributed to phosphate stress (Tai et al., 2009), and similar dynamics could be at play here as well. The Pro99 medium used in this study was calculated to be replete for N, P and other essential inorganic compounds, given cyanobacterial and heterotroph cell abundances and expected cellular requirements (Fagerbakke et al., 1996; Bertilsson et al., 2003). While some nutrient stress could have occurred in our co-cultures, any competition between these two strains was not of a sufficient magnitude to impact Prochlorococcus growth rates or final stationary phase cell density (Figure 1a).
Regulation of transcriptional responses to co-culture
How might all of the transcriptional responses to co-culture described thus far have been regulated? Prochlorococcus has relatively few sigma factors and canonical protein-based transcriptional regulators (Steglich et al., 2008), but a number of these exhibited significant transcriptional enrichment during co-culture—including four sigma factors (PMN2A_0013, PMN2A_1207, PMN2A_1305, PMN2A_1829) as well as multiple potential transcription factors. After 48 h, both the NrdR transcription factor (involved in regulating ribonucleotide reductase expression (Torrents et al., 2007)) and a putative AbrB-family regulator were both transcriptionally enriched in the co-cultures, and a putative GntR-like transcription factor was found to be depleted after 24 h (Supplementary Table S2). Antisense RNAs have an important role in Prochlorococcus transcriptional regulation (Steglich et al., 2008; Voigt et al., 2014), and while they have not yet been characterized in this particular strain, we expect that they likely contributed to at least some of the changes observed.
As noted above, many of the transcriptome changes suggest that co-culture conditions reduced the degree of oxidative stress experienced by Prochlorococcus. Changes in the cellular redox state during co-culture may have been part of the signal sensed by the cell to elicit these responses, either directly transcription factors as described above, or through post-transcriptional regulation. Indeed, the abundance of Rho transcripts, a gene that has an important role in regulating transcriptional termination and contributes to oxidative stress survival in other bacteria (Italiani et al., 2002; Kawamura et al., 2005) decreased after 24 h of co-culture.
Despite the fact that our experiments were conducted under constant illumination conditions, in which cell cycles are not synchronized (Holtzendorff et al., 2001; Zinser et al., 2009; Waldbauer et al., 2012), we observed significant changes in transcript levels for some of the major circadian clock proteins. For example, transcripts encoding LdpA, an input sensor to the circadian clock (Ivleva et al., 2005), were significantly less abundant following 24 h of co-culture, and transcripts for KaiB, one of the central circadian clock proteins, were less abundant after 48 h (Supplementary Table S2). Prochlorococcus has only a partial circadian oscillator that likely needs to be reset daily by sunlight, and so the function of this regulatory oscillator is thought to be influenced by the cellular redox state (Axmann et al., 2009). Thus, we hypothesize that the presence of Alteromonas also impacted Prochlorococcus transcriptional regulation via inputs to the circadian regulation system, whether via impacts on the overall redox state of the cell, changes in the function of the photosynthetic electron transport system, or as a direct or indirect consequence of altering the concentration of reactive oxygen species in the culture.
Response ofAlteromonasto the presence ofProchlorococcus
Thus far we have described only one side of the ‘conversation' between these two strains—that of Prochlorococcus. In these co-culture experiments we used media with no added organics to ensure that Alteromonas would be dependent on fixed carbon released by Prochlorococcus (Bertilsson et al., 2005), as they were in the long-term NATL2A co-cultures before being isolated and maintained alone on simple organic carbon-rich media. At the onset of these experiments, however, the Alteromonas cells were washed and transferred from carbon-rich media into a new environment where, ultimately, the only organic carbon sources were those provided by Prochlorococcus. Although this precluded the possibility of having an Alteromonas-alone control for comparison, we can gain some insights into the heterotroph response by examining transcriptional changes over time following its introduction into the Prochlorococcus culture.
As described above, Alteromonas cell abundance declined following its initial introduction into co-culture with Prochlorococcus, leveled off, and then began to increase after 2 days (Figure 1a). Even if a small amount of organic carbon was provided by lysing cells or media background, this could not have sustained heterotroph growth much beyond the original inoculation density, thus the Alteromonas growth observed must have required consumption of Prochlorococcus exudates. We suspect that this delay in heterotroph growth regrowth resulted from a combination of recovery from stress, acclimation to the new environment, and the amount of time needed for Prochlorococcus to release sufficient photosynthate. Although the heterotroph population size did not increase for multiple days, the RNA data indicate that the population was metabolically active, as the relative amount of heterotroph transcripts in our RNA libraries increased from 3% 2 h after addition to 26% after 12 h of co-culture—even as Prochlorococcus cell abundances continued to increase exponentially over the same time span (Figure 1a; Supplementary Table S1).
An examination of changes in the Alteromonas transcriptome following its introduction into co-culture with Prochlorococcus indicates that these heterotrophs underwent marked changes in their physiology over time. The relative abundance of several hundred Alteromonas transcripts changed over the first 6 h of co-culture, and after 24 h transcripts for ~47% of the genes (1939 out of 4213) in A. macleodii MIT1002 were differentially abundant as compared with the 2 h time point (Table 3). By contrast, transcripts for only 11% of Prochlorococcus genes changed within the co-culture across the same time span. Given the complex heterotroph population dynamics at play here, we focused our analysis on the Alteromonas transcripts that increased in relative abundance during co-culture. Beginning with the 12 h co-culture time point (when the Alteromonas population stopped declining and Prochlorococcus was still growing exponentially), we found 766 proteins that were consistently transcriptionally enriched over the next 36 h (Supplementary Table S3). Although Prochlorococcus exhibited signs of reduced cellular stress in co-culture, the Alteromonas transcriptome suggests that the heterotroph may still have been stressed and potentially nutrient starved for at least the first 2 days of co-culture with Prochlorococcus. For example, multiple stress-responsive transcripts such as those encoding heat shock (hsp20), cold shock (cspG) and phage shock proteins, as well as putative chaperones and genes associated with DNA repair (radC, alkA), all increased in co-culture (Supplementary Table S3). We also saw transcriptional enrichment for stringent starvation protein A, further indicating that the population might have been starved for some nutrient. Perhaps relatedly, there were also consistent increases in transcripts encoding proteins in catabolic processes including amino acid degradation pathways (particularly those for arginine, isoleucine, phenylalanine, tyrosine and valine), degradation of aromatic compounds, and multiple proteases and lipases (Table 4; Supplementary Table S3).
Table 3. Details of changes in the Alteromonas transcriptome during co-culture.
| Time after heterotroph addition (h) | Differentially abundant vs 2h time point | Increased relative abundance vs 12 h time point | Decreased relative abundance vs 12 h time point |
|---|---|---|---|
| 2 | |||
| 4 | 532 | ||
| 6 | 686 | ||
| 12 | 1598 | ||
| 24 | 1939 | 1080 | 711 |
| 48 | 1733 | 1038 | 792 |
Values indicate the number of genes with significantly differentially abundant transcript levels for each indicated pairwise comparison.
Table 4. Alteromonas pathways significantly enriched during co-culture at both the 24 and 48 h co-culture time points as compared with 12 h (see Supplementary Table S3).
| Pathway | Genes with significantly differentially abundant transcript levels | P-value |
|---|---|---|
| Amino acids degradation (arginine, isoleucine, phynylalanine, proline, tyrosine, valine) | astA, metK, putA, MIT1002_0357, MIT1002_0513, MIT1002_0880, MIT1002_0881, MIT1002_0910, MIT1002_0938, MIT1002_1162, MIT1002_1287, MIT1002_2024, MIT1002_2229, MIT1002_2360, MIT1002_2363, MIT1002_2591, MIT1002_2616, MIT1002_2748, MIT1002_2786, MIT1002_2827, MIT1002_2855, MIT1002_3153, MIT1002_3154, MIT1002_3232, MIT1002_3522, MIT1002_3583, MIT1002_3592, MIT1002_3593, MIT1002_3765, MIT1002_3841, MIT1002_4094 | 3.6E-06 |
| Aromatic compounds degradation | dmpN, dmpL, MIT1002_0881, MIT1002_2362, MIT1002_2786, MIT1002_3154, MIT1002_3522, MIT1002_3765, MIT1002_3841 | 4.8E-02 |
| Fatty-acid activation and beta-oxidation | MIT1002_0357, MIT1002_0881, MIT1002_1700, MIT1002_1939, MIT1002_2320, MIT1002_2411, MIT1002_2459, MIT1002_3153, MIT1002_3154, MIT1002_3522, MIT1002_3841 | 4.9E-04 |
| Fatty-acid and lipids degradation | MIT1002_0243, MIT1002_0357, MIT1002_0881, MIT1002_1700, MIT1002_1939, MIT1002_2320, MIT1002_2411, MIT1002_2459, MIT1002_3153, MIT1002_3154, MIT1002_3522, MIT1002_3841 | 1.7E-03 |
| Fatty-acid and lipids biosynthesis | pgpA, pgpB, pssA, MIT1002_0702, MIT1002_1244, MIT1002_1541, MIT1002_1870, MIT1002_2320, MIT1002_2411, MIT1002_2459, MIT1002_3520, MIT1002_3702, MIT1002_3846 | 3.9E-02 |
| Fermentation | ackA, MIT1002_0357, MIT1002_0752, MIT1002_1866, MIT1002_2754, MIT1002_2786, MIT1002_3153, MIT1002_3228, MIT1002_3765 | 1.5E-02 |
| Reactive oxygen species degradation | katG, MIT1002_1531, MIT1002_1576, MIT1002_3838 | 6.2E-02 |
P-value indicates the result of a Fisher's significance test (PathwayTools).
At the same time, the cells also appeared to be remodeling themselves to increase nutrient uptake, with changes in numerous transcripts involved with uptake transport (for compounds such as zinc, choline and tonB transporters for iron uptake) as well as multiple symporters and antiporters (Supplementary Table S3) consistent with a change in their nutrient environment. Multiple transcripts representing phosphorous uptake genes (phoA, phoH, pstS) also consistently increased in relative abundance during co-culture. These results may reflect some aspect of the physiological state of the heterotroph at the outset, but the clear early signs of starvation and the fact that it took many hours before the heterotroph population to begin to grow in co-culture might also indicate that it took Prochlorococcus some time to produce sufficient photosynthate to allow the Alteromonas to expand their population.
Many changes within the heterotroph transcriptome are consistent with the increased expression of pathways needed to support the cell growth observed after 2 days of co-culture (Figure 1a; Table 4). Transcripts involved in anabolic processes such as fatty-acid and lipid biosynthesis consistently increased in abundance (Table 4), as did tRNA synthetases, ATP synthase subunits, as well as transcripts involved in flagellar biosynthesis and central metabolism (for example, pck, ndh, gnd, gapA; Supplementary Table S3). Active cell growth within the population was also suggested by relative increases in transcripts encoding many LSU and SSU ribosomal proteins, as well as DNA replication and cell division proteins (dnaA, scpB, holB, zipA, ftsH, ftsJ; Supplementary Table S3). Transcripts encoding 25 flagellar biosynthesis proteins were also consistently enriched as the population acclimated to co-culture, as were transcripts for RND-family efflux transporters and secreted proteins (Supplementary Table S3). In addition, we also observed a significant increase in the abundance of catalase-peroxidase transcripts (Table 4). This is consistent with the ‘Black Queen' hypothesis (Morris et al., 2012), which posits that the evolutionary loss of the catalase gene in Prochlorococcus was facilitated by the ability of heterotrophs to provide this ‘leaky' function. Although it is unclear which, if any, of these heterotroph transcriptomic responses were specific to co-culture with Prochlorococcus as opposed to changes owing to stresses, growth or simply changes in the nutrient environment, these results support the hypothesis that the chemical conversation in the co-culture was two-sided.
Conclusions and future directions
We have shown that the introduction of a previously sympatric heterotroph to an axenic Prochlorococcus culture can elicit changes in its transcriptome, and presumably affect the downstream physiological processes it mediates. The differences observed in many areas of the Prochlorococcus transcriptome during co-culture—including enrichment of photosystem I transcripts, changes to the electron transport system, ribosomal proteins, transporters and biosynthetic pathways—suggest that co-culture with Alteromonas affected more than just the degree of reactive oxygen stress. In the future it would be interesting to explore the relative impact of oxidative stress reduction vs other impacts of Alteromonas on the relative fitness of Prochlorococcus in co-cultures, measure whether cells do in fact increase carbon fixation rates in co-culture and across different light-limited growth rates. Specific probing of the Prochlorococcus photosystem and electron transfer pathway in co-culture will be necessary to examine whether the changes in PSI transcript abundance resulted in functional changes to photosystem function and light-harvesting capability, and it will be interesting to assess the role of overflow metabolism in generating the extracellular carbon consumed by the heterotroph. Further work to characterize the genes regulated by each transcription factor, and examine the role of small and/or antisense regulatory RNAs in regulating co-culture response, would also shed light onto the regulation of co-culture responses. It is important to recognize that changes in relative transcript abundance within Prochlorococcus does not necessarily always lead to similar changes in protein abundance (Waldbauer et al., 2012). Thus, the changes described here represent a first step toward understanding the impacts of co-culture on Prochlorococcus.
As is true of most laboratory studies of microorganisms in culture, we did not attempt to precisely mimic natural conditions in this work; for example, cell densities were much higher than those in the wild. As the interactions we observed are presumed to be mediated by soluble agents, cell densities could have a role in the outcome—something to be investigated in the future. In addition, our cultures were grown under continuous light to simplify the experimental design, but it would be interesting to explore variations in the outcome across different parts of the day/night light cycle. One of the challenges in designing this type of autotroph–heterotroph co-culture experiment is the nature of the heterotroph control culture, as heterotroph growth was inherently dependent on photosynthate supplied by the autotroph. In future studies, one might compare the transcriptome of the co-cultured heterotroph with the same strain grown on different defined carbon sources to begin to resolve which responses may be specific to Prochlorococcus.
Here we have discussed just one of the myriad possible interacting pairs of Prochlorococcus lineages and the heterotrophs with which they co-occur. The Alteromonas strain used here is a relatively fast-growing (r-selected) heterotroph, and it would be interesting to know how Prochlorococcus might respond to the presence of a more slowly growing (K-selected) heterotroph such as SAR11. As Prochlorococcus and heterotrophs have not, to date, been found to be physically attached to one another (Biller et al., 2015a), we presume that most of the changes observed were triggered by changes in the local environment that, in the wild, would impact the entire community. More generally, these data provide some basis for exploring questions concerning the roles that competitive, commensal and mutualistic interactions play in the physiology of Prochlorococcus, as well as how these types of interactions could have evolved (via ‘Black Queen' dynamics or other mechanisms). No matter how Prochlorococcus responds to any single microbe, given its massive global population, the overall consequences of Prochlorococcus–heterotroph interspecies interactions almost certainly have wide-ranging and multifaceted impacts on marine ecosystems.
Acknowledgments
We thank Jessie Berta-Thompson, Elena Kazamia and Daniel Sher for helpful comments on the manuscript. This work was supported in part by grants from the Gordon and Betty Moore Foundation (GBMF #495.01) and the National Science Foundation (OCE-1356460 and DBI-0424599, the NSF Center for Microbial Oceanography Research and Education). This work was also supported by grants from the Simons Foundation (Grant numbers 337262 to MIT and 329108 to U Hawaii) and is a contribution of the Simons Collaboration on Ocean Processes and Ecology.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Axmann IM, Dühring U, Seeliger L, Arnold A, Vanselow JT, Kramer A et al. (2009). Biochemical evidence for a timing mechanism in Prochlorococcus. J Bacteriol 191: 5342–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam F, Malfatti F. (2007). Microbial structuring of marine ecosystems. Nat Rev Microbiol 5: 782–791. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, Dejongh M, Disz T, Edwards RA et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagby SC, Chisholm SW. (2015). Response of Prochlorococcus to varying CO2:O2 ratios. ISME J 9: 2232–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M et al. (2013). NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41: D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JW, Berube PM, Follett CL, Waterbury JB, Chisholm SW, Delong EF et al. (2014). Closely related phytoplankton species produce similar suites of dissolved organic matter. Front Microbiol 5: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliaev AS, Romine MF, Serres M, Bernstein HC, Linggi BE, Markillie LM et al. (2014). Inference of interactions in cyanobacterial-heterotrophic co-cultures via transcriptome sequencing. ISME J 8: 2243–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berta-Thompson JW. (2015) Prochlorococcus: Life in Light. Ph.D. Thesis, Microbiology Graduate Program, Massachusetts Institute of Technology.
- Bertilsson S, Berglund O, Karl D, Chisholm S. (2003). Elemental composition of marine Prochlorococcus and Synechococcus: Implications for the ecological stoichiometry of the sea. Limnol Oceanogr 48: 1721–1731. [Google Scholar]
- Bertilsson S, Berglund O, Pullin M, Chisholm S. (2005). Release of dissolved organic matter by Prochlorococcus. Vie et Milieu 55: 225–232. [Google Scholar]
- Bertrand EM, Allen AE. (2012). Influence of vitamin B auxotrophy on nitrogen metabolism in eukaryotic phytoplankton. Front Microbiol 3: 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand EM, McCrow JP, Moustafa A, Zheng H, McQuaid JB, Delmont TO et al. (2015). Phytoplankton–bacterial interactions mediate micronutrient colimitation at the coastal Antarctic sea ice edge. Proc Natl Acad Sci 112: 9938–9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube PM, Biller SJ, Kent AG, Berta-Thompson JW, Roggensack SE, Roache-Johnson KH et al. (2015). Physiology and evolution of nitrate acquisition in Prochlorococcus. ISME J 9: 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D, Dufresne A, Vaulot D, Grossman A. (2002). Analysis of the hli gene family in marine and freshwater cyanobacteria. FEMS Microbiol Lett 215: 209–219. [DOI] [PubMed] [Google Scholar]
- Biller SJ, Berube PM, Berta-Thompson JW, Kelly L, Roggensack SE, Awad L et al. (2014). Genomes of diverse isolates of the marine cyanobacterium Prochlorococcus. Sci Data 1: 140034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Berube PM, Lindell D, Chisholm SW. (2015. a). Prochlorococcus: the structure and function of collective diversity. Nat Rev Microbiol 13: 13–27. [DOI] [PubMed] [Google Scholar]
- Biller SJ, Coe A, Martin-Cuadrado A-B, Chisholm SW. (2015. b). Draft genome sequence of Alteromonas macleodii strain MIT1002, isolated from an enrichment culture of the marine Cyanobacterium Prochlorococcus. Genome Announcements 3: e00967–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet S, Webb EA, Panzeca C, Karl DM, Capone DG, Wilhelmy SAS. (2010). Vitamin B12 excretion by cultures of the marine cyanobacteria Crocosphaera and Synechococcus. Limnol Oceanogr 55: 1959–1964. [Google Scholar]
- Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA et al. (2014). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 42: D459–D471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm SW, Frankel SL, Goericke R, Olson RJ, Palenik B, Waterbury JB et al. (1992). Prochlorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b. Arch Microbiol 157: 297–300. [Google Scholar]
- Coleman ML, Chisholm SW. (2007). Code and context: Prochlorococcus as a model for cross-scale biology. Trends Microbiol 15: 398–407. [DOI] [PubMed] [Google Scholar]
- Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438: 90–93. [DOI] [PubMed] [Google Scholar]
- Fagerbakke KM, Heldal M, Norland S. (1996). Content of carbon, nitrogen, oxygen, sulfur and phosphorus in native aquatic and cultured bacteria. Aquat Microbial Ecol 10: 15–27. [Google Scholar]
- Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N et al. (2013). Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci 110: 9824–9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk C, Vermaas W. (1999). A cyanobacterial gene family coding for single-helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry 38: 9397–9404. [DOI] [PubMed] [Google Scholar]
- Giannoukos G, Ciulla DM, Huang K, Haas BJ, Izard J, Levin JZ et al. (2012). Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol 13: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez JC, Banerjee RV, Huang S, Sumner JS, Matthews RG. (1992). Comparison of cobalamin-independent and cobalamin-dependent methionine synthases from Escherichia coli: two solutions to the same chemical problem. Biochemistry 31: 6045–6056. [DOI] [PubMed] [Google Scholar]
- He Q, Dolganov N, Bjorkman O, Grossman AR. (2001). The high light-inducible polypeptides in Synechocystis PCC6803. Expression and function in high light. J Biol Chem 276: 306–314. [DOI] [PubMed] [Google Scholar]
- Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG. (2011). Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol Biol Evol 28: 2921–2933. [DOI] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB. (2010). Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8: 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzendorff J, Partensky F, Jacquet S, Bruyant F, Marie D, Garczarek L et al. (2001). Diel expression of cell cycle-related genes in synchronized cultures of Prochlorococcus sp. strain PCC 9511. J Bacteriol 183: 915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiani VCS, Zuleta LFG, Marques MV. (2002). The transcription termination factor Rho is required for oxidative stress survival in Caulobacter crescentus. Mol Microbiol 44: 181–194. [DOI] [PubMed] [Google Scholar]
- Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. (2005). LdpA: a component of the circadian clock senses redox state of the cell. EMBO J 24: 1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Paley SM, Krummenacker M, Latendresse M, Dale JM, Lee TJ et al. (2010). Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinformatics 11: 40–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura N, Kurokawa K, Ito T, Hamamoto H, Koyama H, Kaito C et al. (2005). Participation of Rho-dependent transcription termination in oxidative stress sensitivity caused by an rpoB mutation. Genes Cells 10: 477–487. [DOI] [PubMed] [Google Scholar]
- Kazamia E, Czesnick H, Nguyen TTV, Croft MT, Sherwood E, Sasso S et al. (2012). Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ Microbiol 14: 1466–1476. [DOI] [PubMed] [Google Scholar]
- Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S et al. (2007). Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet 3: e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell D, Jaffe JD, Coleman ML, Futschik ME, Axmann IM, Rector T et al. (2007). Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 449: 83–86. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfatti F, Azam F. (2009). Atomic force microscopy reveals microscale networks and possible symbioses among pelagic marine bacteria. Aquat Microbial Ecol 58: 1–14. [Google Scholar]
- Malmstrom RR, Coe A, Kettler GC, Martiny AC, Frias-Lopez J, Zinser ER et al. (2010). Temporal dynamics of Prochlorococcus ecotypes in the Atlantic and Pacific oceans. ISME J 4: 1252–1264. [DOI] [PubMed] [Google Scholar]
- Mella-Flores D, Six C, Ratin M, Partensky F, Boutte C, Le Corguillé G et al. (2012). Prochlorococcus and Synechococcus have evolved different adaptive mechanisms to cope with light and UV stress. Front Microbiol 3: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L, Coe A, Zinser ER, Saito M, Sullivan M, Lindell D et al. (2007). Culturing the marine cyanobacterium Prochlorococcus. Limnol Oceanogr 5: 353–362. [Google Scholar]
- Morris JJ, Johnson ZI, Szul MJ, Keller M, Zinser ER. (2011). Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean's surface. PLoS One 6: e16805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. (2008). Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by ‘helper' heterotrophic bacteria. Appl Environ Microbiol 74: 4530–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JJ, Lenski RE, Zinser ER. (2012). The Black Queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3: e00036–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivals I, Personnaz L, Taing L, Potier M-C. (2007). Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23: 401–407. [DOI] [PubMed] [Google Scholar]
- Saito M, Moffett J, Chisholm S, Waterbury J. (2002). Cobalt limitation and uptake in Prochlorococcus. Limnol Oceanogr 47: 1629–1636. [Google Scholar]
- Sañudo-Wilhelmy SA, Gómez-Consarnau L, Suffridge C, Webb EA. (2013). The role of B vitamins in marine biogeochemistry. Annu Rev Marine Sci 6: 339–367. [DOI] [PubMed] [Google Scholar]
- Scanlan DJ, Hess WR, Partensky F, Newman J, Vaulot D. (1996). High degree of genetic variation in Prochlorococcus (Prochlorophyta) revealed by RFLP analysis. Eur J Phycol 31: 1–9. [Google Scholar]
- Sher D, Thompson JW, Kashtan N, Croal L, Chisholm SW. (2011). Response of Prochlorococcus ecotypes to co-culture with diverse marine bacteria. ISME J 5: 1125–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich C, Futschik M, Rector T, Steen R, Chisholm SW. (2006). Genome-wide analysis of light sensing in Prochlorococcus. J Bacteriol 188: 7796–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich C, Futschik ME, Lindell D, Voss B, Chisholm SW, Hess WR. (2008). The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet 4: e1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai V, Paulsen IT, Phillippy K, Johnson DA, Palenik B. (2009). Whole-genome microarray analyses of Synechococcus-Vibrio interactions. Environ Microbiol 11: 2698–2709. [DOI] [PubMed] [Google Scholar]
- Thompson AW, Huang K, Saito MA, Chisholm SW. (2011). Transcriptome response of high- and low-light-adapted Prochlorococcus strains to changing iron availability. ISME J 5: 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolonen AC, Aach J, Lindell D, Johnson ZI, Rector T, Steen R et al. (2006). Global gene expression of Prochlorococcus ecotypes in response to changes in nitrogen availability. Mol Syst Biol 2: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents E, Grinberg I, Gorovitz-Harris B, Lundström H, Borovok I, Aharonowitz Y et al. (2007). NrdR controls differential expression of the Escherichia coli ribonucleotide reductase genes. J Bacteriol 189: 5012–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt K, Sharma CM, Mitschke J, Lambrecht SJ, VoB B, Hess WR et al. (2014). Comparative transcriptomics of two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity. ISME J 8: 2056–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldbauer JR, Rodrigue S, Coleman ML, Chisholm SW. (2012). Transcriptome and proteome dynamics of a light-dark synchronized bacterial cell cycle. PLoS One 7: e43432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Cho Y-J, Won S, Lee J-E, Jin Yu H, Kim S et al. (2011). Duplex-specific nuclease efficiently removes rRNA for prokaryotic RNA-seq. Nucleic Acids Res 39: e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser ER, Lindell D, Johnson ZI, Futschik ME, Steglich C, Coleman ML et al. (2009). Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS One 4: e5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorz J, Allanach J, Murphy C, Roodvoets M, Campbell D, Cockshutt A. (2015). The RUBISCO to photosystem II ratio limits the maximum photosynthetic rate in picocyanobacteria. Life 5: 403–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.