Abstract
The ability to navigate through space involves complex interactions between multiple brain systems. Although it is clear that spatial navigation is impaired during aging, the networks responsible for these altered behaviors are not well understood. Here, we used a within-subject design and [18F]FDG-microPET to capture whole-brain activation patterns in four distinct spatial behaviors from young and aged rhesus macaques: constrained space (CAGE), head-restrained passive locomotion (CHAIR), constrained locomotion in space (TREADMILL), and unconstrained locomotion (WALK). The results reveal consistent networks activated by these behavior conditions that were similar across age. For the young animals, however, the coactivity patterns were distinct between conditions, whereas older animals tended to engage the same networks in each condition. The combined observations of less differentiated networks between distinct behaviors and alterations in functional connections between targeted regions in aging suggest changes in network dynamics as one source of age-related deficits in spatial cognition.
SIGNIFICANCE STATEMENT We report how whole-brain networks are involved in spatial navigation behaviors and how normal aging alters these network patterns in nonhuman primates. This is the first study to examine whole-brain network activity in young or old nonhuman primates while they actively or passively traversed an environment. The strength of this study resides in our ability to identify and differentiate whole-brain networks associated with specific navigational behaviors within the same nonhuman primate and to compare how these networks change with age. The use of high-resolution PET (microPET) to capture brain activity of real-world behaviors adds significantly to our understanding of how active circuits critical for navigation are affected by aging.
Keywords: aging, brain circuits, spatial cognition
Introduction
The ability to navigate environments effectively involves orchestrated interactions among many brain regions. Despite the widely varying ages at which an animal is considered to be “old,” reduced spatial navigation accuracy has been reported in all aged mammals studied (Barnes, 1979; Rapp et al., 1997; Moffat et al., 2001; Samson and Barnes, 2013). Therefore, developing an understanding of how advanced age affects neural circuits that support spatial navigation is imperative for promoting positive health outcomes in older populations. Electrophysiological and MRI methods used to examine this question in humans and rodents each have distinct strengths and drawbacks with respect to identifying which networks are responsible for altered spatial cognition with age (Barnes et al., 1997; Moffat and Resnick, 2002; Wilson et al., 2005; Wiener et al., 2013). For example, high-density ensemble recording methods applied in rodents can be conducted during unrestrained behavioral states and have outstanding temporal resolution (Wilson and McNaughton, 1993; Rosenzweig et al., 2003), but the number of brain regions that can be recorded simultaneously is limited compared with the broader brain surveys possible with noninvasive MRI methods (Poser et al., 2010; Goense et al., 2010). Virtual navigation paradigms that can be administered in the scanner in humans, however, do not allow free locomotion.
Both high-density rodent recording studies and virtual navigation paradigms in humans have implicated the hippocampus and other temporal lobe regions as contributors to spatial memory and navigation impairments in aging (Barnes et al., 1997; Moffat et al., 2001; Moffat and Resnick, 2002; Wilson et al., 2005; Konishi and Bohbot, 2013; Wiener et al., 2013). Studies in humans have also identified a number of other cortical and subcortical regions, in addition to the hippocampus, that are active during virtual navigation paradigms (Aguirre et al., 1996; Maguire et al., 2000; Hartley et al., 2003; Bohbot et al., 2007; Ekstrom et al., 2009). Although the possibility exists that participants can implement different behavioral strategies to solve a given problem (Head and Isom, 2010), older adults have been found to show reduced functional activation in fMRI studies during virtual environment navigation (Moffat et al., 2006). The present study was designed explicitly to examine brain activity after different forms of locomotion through space, including completely free movement, in a nonhuman primate model of normal aging.
FDG-microPET was used to examine metabolic activity induced by unrestrained and controlled locomotion behaviors emitted immediately before scanning. For the purposes of the present experiment, we refer to “navigation” as the state in which the monkey is free to forage for food reward with no restraint in locomotion other than the boundaries of the enclosure with full optic flow and vestibular components engaged. Whole-brain activation patterns evoked by this type of navigation behavior and three control behaviors are examined within the same individual young and old monkeys as follows: unconstrained locomotion (WALK), head-restrained passive locomotion (CHAIR), constrained locomotion in space (TREADMILL), and constrained space (CAGE) (Fig. 1). This allowed us to address two fundamental hypotheses: (1) that distinct networks will be engaged and activated differentially during different forms of locomotion and (2) that age will alter the composition of these networks across the different behavioral conditions.
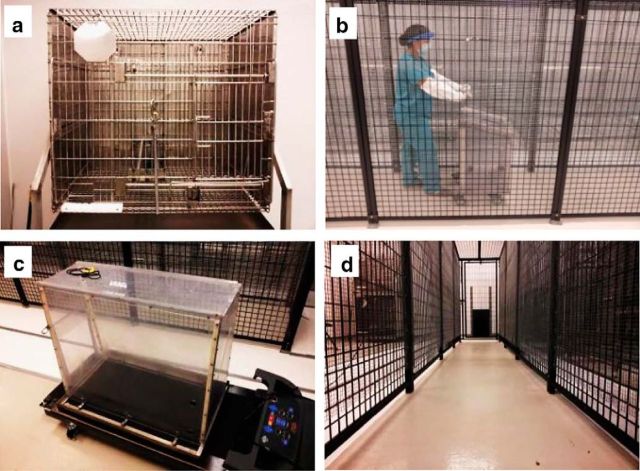
Figure 1.
Photographs illustrating the four experimental treatments in our constrained, restrained, and unconstrained locomotion behaviors. These included constrained space (a; CAGE),passive locomotion through the 6 m enclosure in a nonhuman primate restraint chair, with head restraint (b; CHAIR), constrained locomotion in space (c; TREADMILL), and unconstrained locomotion in the 6 m enclosure (d; WALK).
Materials and Methods
Subjects.
Three young and three aged (Table 1) rhesus macaques (Macaca mulatta) participated in our real-world freely moving conditions and restrained and constrained locomotion conditions (equivalent human age range ∼24–84 years). The behavioral conditions were followed by microPET scans to determine differential activation of specific areas of the brain under different types of movement protocols, as described in detail below. Five of the monkeys were male. Four monkeys (two young and two aged) were surgically implanted with a recording chamber and a head post (Crist Instruments) in preparation for future electrophysiological studies (for details, see Skaggs et al., 2007) and two monkeys were fitted with a thermoplastic mask (Civco) to restrain head movement in the primate chair, passive movement condition. Each monkey was opposite-sex paired housed in an environmentally controlled room and maintained on a 12 h light/dark cycle at the California National Primate Research Center (CNPRC) at the University of California–Davis. Each monkey received daily monkey chow (Lab Diet #5047; PMI Nutrition International); ad libitum water, supplemental fruit, nuts, and vegetables; and weekly enrichment that varied from puzzle ball feeders to hour-long visits to a large exercise arena. All experimental procedures were performed in accordance with the National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee at the University of California–Davis.
Table 1.
Demographics and behavioral performance characteristics in WALK condition
| Animal | Age | Sex | No. of laps | Time into run (min:sec) | Time at last lap | Time out of run | Mean GM* |
|---|---|---|---|---|---|---|---|
| 35481 | 8 y 11 mo | M | 54 | 4:03 | 20:14 | 20:51 | 111.04 |
| 35765 | 8 y 1 mo | M | 34 | 2:02 | 19:57 | 20:12 | 119.58 |
| 31299 | 12 y 8 mo | M | 21 | 2:38 | 20:11 | 20:17 | 118.38 |
| 23938 | 25 y 5 mo | M | 27 | 3:00 | 20:42 | 21:10 | 118.21 |
| 22119 | 27 y | M | 27 | 2:33 | 20:31 | 20:31 | 108.54 |
| 21899 | 27 y 9 mo | F | 18 | 2:29 | 20:09 | 20:11 | 114.75 |
*Mean GM = global mean metabolism of gray matter from each monkey during the WALK condition.
Behavioral apparati and behavioral tasks.
There were four behavioral conditions (Fig. 1): (1) a standard 0.864 m × 0.838 m × 0.711 m primate cage to constrain space (CAGE); (2) a nonhuman primate restraint chair in which the animal could be head restrained and moved passively (CHAIR); (3) a 0.457 m × 0.965 m × 0.838 m modified treadmill to constrain locomotion in space (TREADMILL); and (4) a 6.096 m × 0.9144 m × 1.829 m enclosure for larger-scale unconstrained locomotion (WALK). The primate cage did not eliminate locomotion, but constrained it within a more limited space than the 6 m enclosure. The timing and numbers of rewards were matched to the other conditions and delivered at the front of the cage. The treadmill was modified so that the deck was enclosed with clear polycarbonate. Feeding ports were positioned on the front of the treadmill enclosure for reward administration so that a similar number of rewards could be delivered as in the WALK, CHAIR, and CAGE conditions. The primate restraint chair was made of polycarbonate and was used to move the monkey passively through the long enclosure environment to engage optic flow and to match the number of laps traversed and rewards delivered while eliminating locomotion. The long enclosure was made of painted steel and was lined with 12.7 mm grid wire. It had two doors, one at each end, which served as entry points into the long enclosure, and formed the primary unconstrained locomotion condition with normal optic flow, locomotion, and reward delivery (two rewards per lap, one at each end) to compare with the other, more constrained or restrained conditions.
Before testing, each monkey was trained until it participated consistently for at least 20 min without any coaxing (other than food rewards) and did not display any apparent sign of distress. The first treatment that each monkey participated in was the freely moving condition in the 6 m enclosure (WALK). The amount of time allotted per testing session was 20 min. The total time, lap number, number of rewards, reward acceptance (i.e., whether taken directly by mouth or by which hand or if the reward was refused), and any notable behaviors were manually recorded during this testing session to serve as reference data to be matched across treatment conditions for any given animal. The experiment was designed this way to keep as many factors constant across the three active conditions to best capture the major variables of interest. Following behavior in the long enclosure, the remaining three treatment conditions (CAGE, CHAIR, and TREADMILL) were conducted in a pseudorandom order. In addition, before testing, each monkey was trained to present their arm and acclimated to intravenous injection to adapt them to accept radiotracer administration. This occurred across multiple sessions during the light cycle to decrease the amount of agitation each monkey experienced during radiotracer administration (Holschneider and Maarek, 2008).
MRI scanning.
T1-weighted structural MRI scans were acquired with a 1.5 T Genesis Signa Model scanner (General Electric) at the University of California–Davis Imaging Research Center for each monkey before participation in the current microPET study (for details, see Alexander et al., 2008). Under isoflurane gas anesthesia, ∼80 1.0-mm-thick images were acquired using a T1-weighted spoiled gradient (SPGR) pulse sequence (repetition time = 22 ms; echo time = 7.9 ms; number of excitations = 3; FOV = 16 cm; matrix = 256 × 256).
microPET scanning.
A transmission scan was collected while animals underwent WALK behavioral training to serve as a positional reference and for the 3D reprojection attenuation correction algorithm for the emission scans that were acquired after behavioral testing. This scan was obtained before any of the emission scans were obtained and during the period that the animals were being trained to walk continuously on the 6 m track and on the treadmill. Alignment of the transmission and emission scans was automated and inspected visually for quality assurance. Four emission scans were collected from each animal following the intravenous infusion of [18F]FDG (PETNET Radiopharmaceuticals). The following protocol was used, which only differed by which treatment condition was administered, before each emission scan. On the day of the emission scan, the monkey was transported to our testing room, placed into a holding cage, and presented its arm to be shaved, cleaned, and catheterized, at which time a blood sample was drawn to measure blood glucose level. Time 0 for each treatment condition began with the intravenous injection of the [18F]FDG radiotracer (0.5 mCi/kg), followed by a sterile saline flush. The monkey then participated in one of the four treatment conditions. For the CAGE condition, the monkey remained in the holding cage for 20 min. For the other three conditions, the monkey was returned to the holding cage after a 20 min behavior session and anesthetized with an intramuscular injection of ketamine (10 mg/kg). Upon sedation, each monkey was immediately transferred to the microPET imaging facility at the CNPRC and placed in a supine position on the microPET scanner bed (Model P4; Siemens) and immediately intubated with an endotracheal cannula. Anesthesia level was maintained by isoflurane gas (1.5–3%), whereas heart rate, respiratory rate, blood oxygenation, and CO2 levels were monitored by the veterinary staff. When vital signs were stable, the animal was then secured to the scanner bed with either a custom PVC head holder that secured a Crist Instrument titanium head post (two young and two old monkeys) or by an individualized thermoplastic device to position and orient the monkey's head across emission scans (one young and one old monkey). The P4 scanner has a 22 cm animal port, a 19 cm transaxial FOV and a 7.8 cm axial FOV. After laser guided head centering, each monkey was moved into the microPET scanner and a 1 min scan was used to ensure that the entire brain was within the field of view and to check for position and orientation alignment to the baseline transmission scan. A 60 min static emission scan was started on average 60.3 min (range 60–63) after injection to measure [18F]FDG metabolism throughout the brain. After each emission scan, each monkey recovered in a designated room with its normal pair-housed cage mate and was returned to its home cage within 24 h after clearance of the radiotracer. The emission scan interval between scanning conditions was set to a minimum of 7 d (range 7–41).
Image preprocessing.
The methods applied to the MRI and microPET images are adaptations of voxel-based methods from previous work in nonhuman primates and humans to perform partial least-squares (PLS) analysis (Wisco et al., 2008; e.g., Alexander et al., 2008). Image processing was performed by 3D slicer (Slicer 4.3.2, www.slicer.org/slicer4), and MRI and microPET datasets were imported as DICOM files. Each monkey's T1-weighted SPGR MRI was preprocessed following voxel-based morphometric methods to create a within study template (for details, see Alexander et al., 2008). The index number before each processing step below corresponds to those illustrated in Figure 2. Each MRI was processed as follows: (1) manually transformed along the anterior commissure–posterior commissure line and zeroed on the posterior commissure; (2) inhomogeneity corrected; (3) manually skull stripped; and (4) one monkey was selected to serve as the initial template for future registrations. From here, each skull-stripped MRI was processed as follows: (5) histogram equalized to match the template MRI; (6) resampled to 0.5 mm3 voxel size; (7) segmented into gray matter and white matter probability maps by the EM segmenter module (Pohl et al., 2007; Fedorov et al., 2011); and (8) registered to the template MRI linear and nonlinear B-spline transformation. The registered probability maps were then: (9) averaged together to create the within study template. After the creation of the within-study MRI template: (10) each resampled native scan was then registered to the within study template. Each microPET scan was then: (11) isotropically cropped and resampled at 0.5 mm3 voxels; (12) spatially coregistered to their respective SPGR MRI by a linear transformation; and (13) skull stripped. Each skull-stripped microPET scan was then intensity normalized by: (14) the extracted average intensity of the white matter of each microPET scan by applying each monkey's MRI segmented white matter probability map as an ROI; (15) ratio normalized relative to the white matter to account for differences in injected activity across conditions within and between animals (Borghammer et al., 2008) and then multiplied by 100 to scale as a percentage, which was performed through the python extension of Slicer; (16) spatially normalized to the within-study template by applying the same transformations that were created during the MRI-to-template phase of the pipeline; and, finally, 917) spatially smoothed with a Gaussian filter (FWHM = 6 mm; Alexander et al., 2008). Quality assurance was performed at each phase of the pipeline. Each scan was then converted and saved in Analyze floating point format and subsequently exported into Matlab for PLS analysis. For independent MRI analysis, cortical gray matter and cerebellum segmentation label maps were used to extract ratio normalized gray matter and cerebellum values for independent MRI data analysis. This was performed in 3D slicer label statistics module and exported for statistical analysis in SPSS version 20.
Figure 2.
Highlighted steps of the image-processing pipeline for the MRI and microPET scans. These steps included: (1–6) MRI images were cropped, zeroed along the anterior/posterior commissure line, and manually skull stripped; (7–8) segmented into gray and white matter probability maps; and then (9–10) averaged together to create a within study template. PET images: (11–13) followed similar preprocessing steps before registration to each animals respective MRI; (14–15) intensity normalized relative to the white matter; (16) spatially normalized to the within study template; and then (17) spatially smoothed with a 6 mm Gaussian filter.
Statistical analysis.
PLS correlation analysis (McIntosh et al., 1996; for computations, see Krishnan et al., 2011) was used to identify patterns of FDG metabolism that were related to the CAGE, CHAIR, TREADMILL, and WALK treatment conditions and to determine the functional connectivity of select regions that contribute to the different locomotion behaviors. PLS correlation was performed with the freely available PLSgui (http://www.rotman-baycrest.on.ca) that is run in Matlab R2012b (The MathWorks). PLS correlation is designed to maximize the covariance between two data matrices in single-subject and group neuroimaging designs (McIntosh and Lobaugh, 2004) and has been used previously on fMRI and PET modalities to differentiate patterns of brain activity that are associated with different treatment conditions (task-PLS) or to determine the functional connectivity (seed-PLS) of an ROI (McIntosh et al., 1996, 1999; Cabeza et al., 1997; Grady et al., 2012). In the context of the present study, one data matrix reflected brain activity in young and aged monkeys (two submatrices, focusing on the age variable). The second data matrix, however, depended on the type of PLS analysis performed. For task-PLS, the second matrix represented voxel activity from the PET data from corresponding treatment conditions (four submatrices). For behavior-PLS, the second data matrix represented quantified behavioral data such as number of laps and rewards for behavior-PLS. For seed-PLS, the matrix reflected activity from a selected set of voxels (5 mm seed).
Depending on the type of PLS analysis conducted, the next step is to derive voxelwise condition means. Voxelwise grand means are then calculated across the different treatment conditions for task PLS or a correlation matrix is derived for the seed-PLS. The original data matrices are then combined and a mean-centered data matrix is created by subtracting either the condition means or the grand mean depending on whether the question pertains to treatment condition or age group differences. The mean-centered data matrix or correlation matrix then undergoes singular value decomposition to form latent variables, which reveal the covariance between brain activity and the treatment conditions (i.e., positive latent variable scores indicate a positive linear relationship, whereas negative latent variable scores indicate an inverse linear relationship). The total number of possible latent variables is limited by the number of groups and treatments—in this case, two age groups of animals and four treatment conditions yielded eight possible latent variables—whereas the number of significant latent variables is determined by the rank of the data matrix. The proportion of variance accounted for by each latent variable is identified and the statistical significance (p < 0.05) of each is determined by using a permutation test without sampling replacement (N = 500 iterations). To illustrate and summarize the pattern of activity found in each age group and across the four treatment conditions, mean brain score plots were created by summing the latent brain variable scores in each monkey across the different treatment conditions for every significant latent variable.
To determine significant differences in brain activity across the treatment conditions in the young and aged monkey groups, a bootstrap resampling procedure (n = 6 resamples) was used to create 95% confidence intervals. Non-overlapping confidence intervals between age groups and treatment conditions are used to indicate statistical differences. To identify brain activity associated with each latent variable, all voxels in which the bootstrap ratio (salience/SE) was greater than ±3 (p = 0.005) were considered to be a statistically significant contributor to the mean brain score for each latent variable. A minimum cluster size of 200 contiguous voxels with a minimum peak-to-peak distance of 5 mm was used to identify stable clusters. Latent variable image data are illustrated as horizontal slice by slice and lateral 3D cortical-surface maps as bootstrap ratio scores that met the above criteria. Differences in activity between groups were further examined by using the identified network patterns as ROIs. This was accomplished by exporting the network pattern ROIs as label maps into 3D slicer. The mean FDG metabolism values over these ROIs were extracted from each PET image. A two-group ANOVA [(young vs older) × two network (positive or negative salience group scores) × four condition (CAGE, CHAIR, TREADMILL, and WALK)] was performed to detect differences (p < 0.05) in FDG metabolism between groups as a function of network and condition in SPSS version 20.
Results
Patterns of activity across treatment conditions
Variants of PLS analysis were used to identify whole-brain patterns of FDG metabolism associated with the four treatment conditions (Fig. 1) in young and old monkeys. First, we used task-PLS to identify covarying patterns of activity across the four treatment conditions in both age groups. This revealed two significant latent variables (LVs) or coactivation patterns of FDG metabolism (LV1 and LV2) that accounted for 35% and 30.9% of the variance in the data (p < 0.05), respectively.
The first pattern (LV1; Fig. 3, Table 2) distinguished areas of coactivation generated by active and passive locomotion through large-scale space (CHAIR and WALK) from movement in constrained space (CAGE and TREADMILL). Although regions coactivated in these conditions were similar, the differentiation between these behaviors was weaker in the older monkeys (Fig. 3a). Notable regions that showed greater coactivity during the CHAIR and WALK condition included the occipital lobes, superior temporal gyrus (STG), orbitofrontal cortex, posterior cingulate cortex, retrosplenium, right posterior hippocampus, right entorhinal cortex, left posterior perirhinal cortex, parahippocampal cortex, temporal area E (TE), left amygdala, body of the caudate nucleus, anterior thalamus, and the hypothalamus. The CAGE and TREADMILL conditions were associated with greater FDG coactivity patterns in a different set of regions, including posterior areas of the frontal and lateral areas of the parietal lobes, right anterior perirhinal cortex, ventral cerebellum, head of caudate nucleus, and the putamen.
Figure 3.
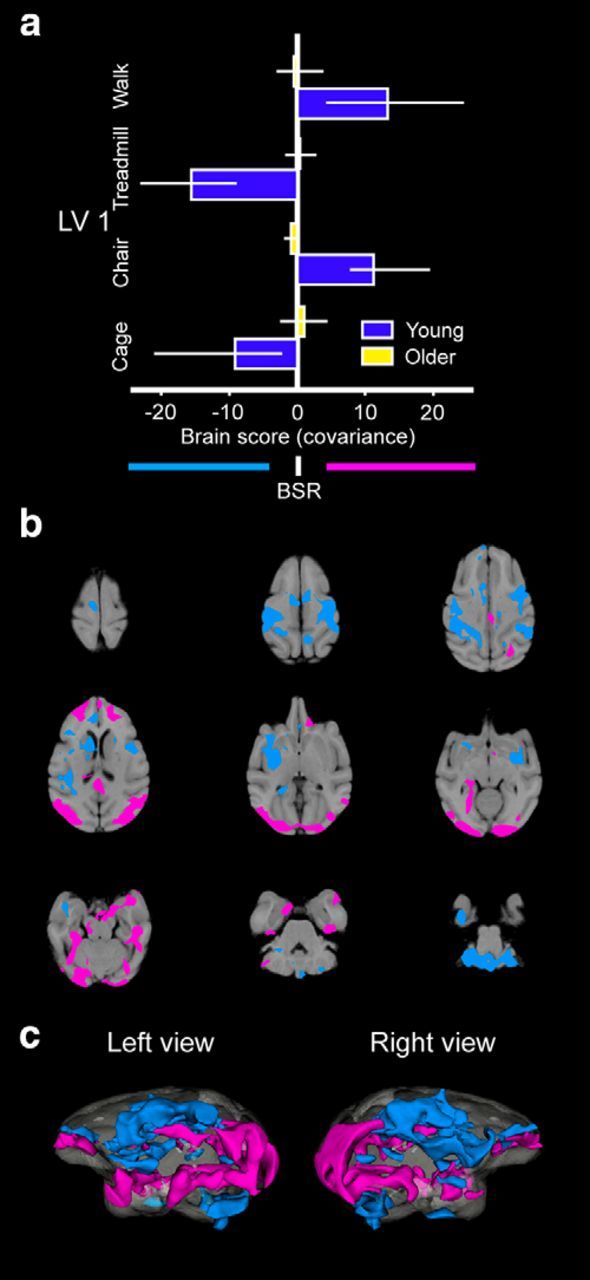
Task-PLS results for LV 1 as a function of the different treatment conditions and age. Mean-centered brain score plots reveal a pattern of FDG metabolism that differentiated the CHAIR and WALK conditions from the CAGE and TREADMILL conditions (a). Error bars represent 95% confidence intervals for the calculated patterns using a bootstrap procedure. Nonoverlapping confidence intervals indicate differences between the conditions and groups of monkeys. Patterns of whole-brain FDG metabolism on horizontal slice by slice 2D sections are shown (b) superimposed on top of the within-study template and reconstructed 3D metabolism maps (c) illustrate the areas associated with this pattern. Colors represent values for positive (magenta) and negative (cyan) bootstrap ratio (BSR) values that are ≥±3 (p < 0.005), reflecting the distinct networks associated with these specific behaviors.
Table 2.
Brain regions and clusters associated with latent variable 1 (LV 1)
| Region | Hemisphere | X | Y | Z | BSR |
|---|---|---|---|---|---|
| V1 | Right | 11 | 1 | −30.5 | 133.323 |
| Cerebellum | Left | −14 | −11 | 0.25 | 15.1739 |
| V3 | Right | 15.5 | 11 | −22 | 100.1645 |
| V3A | Left | −19 | 10 | −18.5 | 105.1515 |
| Medial parietal | Left | −4.5 | 9.5 | −11.5 | 17.2297 |
| TEO | Left | −22.5 | −5.5 | −9.5 | 4.461 |
| STG | Left | −24 | −5.5 | −0.5 | 5.682 |
| Pulvinar | Right | 6 | 7 | −2 | 28.3884 |
| Post. Hippocampus | Right | 14 | −5.5 | 3 | 7.316 |
| Parahippocampal | Left | −18.5 | −10.5 | 0.5 | 145.5612 |
| Entorhinal | Right | 7.0 | −12 | 11 | 5.731 |
| Retrosplenium | Left | −1.5 | 8.5 | −8 | 6.781 |
| Globus pallidus | Left | −9 | −5 | 11.5 | 89.3022 |
| Caudate | Right | 1 | 5.5 | 12 | 25.3225 |
| Amygdala | Right | 7.5 | −11.5 | 12.5 | 9.8296 |
| TAa | Right | 23.5 | −10.5 | 17.5 | 27.3766 |
| Orb. frontal gyrus | Left | −8 | 6.5 | 31.5 | 79.1194 |
| 47 | Right | 8 | 8 | 39 | 31.6171 |
| 32 | Left | 0 | 8.5 | 40.5 | 21.6735 |
| Posterior cingulate | Left | −2.0 | 9.5 | −7 | 13.837 |
| Hypothalamus | Left | −2.0 | −5.5 | 8 | 5.247 |
| Caudate | Left | −6.5 | 6.0 | 13.5 | 16.311 |
| Cerebellum | Left | −1.5 | 1 | −21 | −6.2437 |
| Cerebellum | Right | 8.5 | −17 | −15.5 | −189.435 |
| Medial parietal | Left | −4.5 | 16.5 | 12 | −36.5773 |
| V1 | Right | 8 | 2.5 | 8 | −23.9021 |
| PECg | Left | −6 | 15 | −3.5 | −31.4272 |
| S2 | Right | 16.5 | 7.5 | −2.5 | −139.946 |
| Area 2 | Left | −18.5 | 15.5 | 3 | −90.3917 |
| Perirhinal | Right | 16.5 | −17.5 | 8.5 | −16.5628 |
| Globus pallidus | Left | −12.5 | −4 | 10 | −39.6587 |
| Cingulate | Right | 5.5 | 14.5 | 11 | −164.28 |
| Parainsular | Right | 20.5 | −6 | 12 | −30.1334 |
| Putamen | Right | 12.5 | 1 | 16.5 | −121.156 |
| Caudate | Right | 5 | 5 | 22 | −66.2413 |
| Area 32 | Right | 4 | 9.5 | 33 | −46.4201 |
X, Y, Z, Spatial coordinates in relation to the posterior commissure; BSR, bootstrap ratio values greater than our criterion (BSR > 3, p = 0.005).
To explore the observation that between-condition FDG coactivity patterns appeared more distinct in the young monkeys (Fig. 3a), we used task-PLS weighted for a group interaction (mean centering type 1 method) to determine whether the spatial patterns were different between age groups. There was no significant difference in brain areas activated between age groups (p > 0.10), suggesting that the age difference is in the degree to which the old monkeys engage the same areas that were found to be active in the young monkeys. We then used a variant of PLS (behavior-PLS) to determine whether FDG metabolism was influenced by performance variables (i.e., the number of laps in the linear track). There was no significant group difference (p > 0.10), suggesting that these performance variables cannot explain the age difference. To support this observation, we also extracted the global mean metabolism of the gray matter from each monkey during the WALK condition and compared these values with the number of laps that each monkey ran (Table 1). There was no significant correlation between laps completed and overall metabolic activity (p > 0.05). Therefore, the first pattern distinguished a network coactive in passive or active locomotion through large-scale space from a network coactive in movement in constrained space, neither of which was as well differentiated in old monkeys.
The second pattern (LV2; Fig. 4, Table 3) distinguished areas of coactivation generated by active locomotion (TREADMILL and WALK) from those generated during passive or limited movement (CAGE and CHAIR). Again, the regions coactivated in these conditions were similar, but the differentiation between these behaviors was weaker in the older monkeys (Fig. 4a). Regions that showed coactive FDG metabolism during the TREADMILL and WALK conditions included large clusters in the occipital lobe, MT, medial superior temporal area (MST), retrosplenium, posterior cingulate cortex, dorsal cerebellum, central sulcus, right posterior hippocampus, right colliculi, left pulvinar, thalamus, and widespread regions throughout the temporal lobe. The CAGE and CHAIR conditions were associated with coactivation patterns in a different set of regions including the ventral cerebellum, STG, TE, perirhinal cortex, temporal pole, intraparietal sulcus, anterior thalamus, hypothalamus, left middle and anterior hippocampus, amygdala, and medial orbitofrontal areas. Therefore, the second pattern distinguished a network coactive during free locomotion from a network coactive in locomotor-constrained behaviors, which was not as well differentiated in old monkeys.
Figure 4.
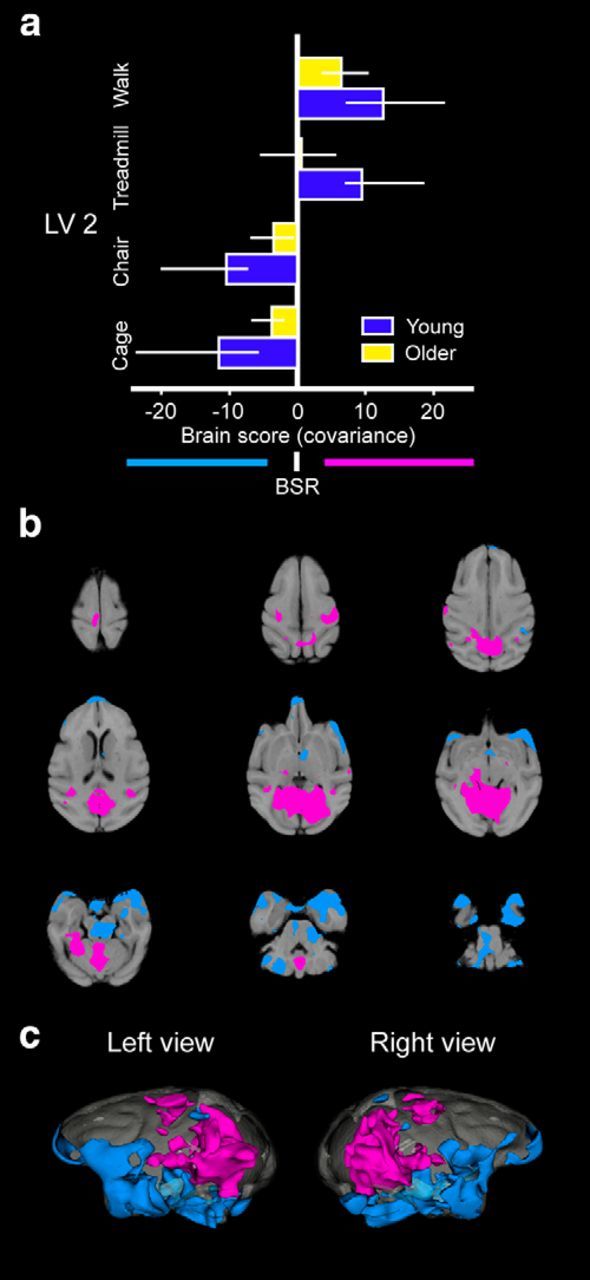
Task-PLS results for LV2 as a function of the different treatment conditions and age. Mean-centered brain score plots reveal a pattern of FDG metabolism that differentiated the TREADMILL and WALK conditions from the CAGE and CHAIR conditions (a). Patterns of whole-brain FDG metabolism on horizontal slice by slice 2D sections (b) superimposed on top of the within study template and reconstructed 3D metabolism maps (c) illustrate the areas associated with this pattern. Conventions are the same as in Figure 3.
Table 3.
Brain regions and clusters associated with latent variable 2 (LV 2)
| Region | Hemisphere | X | Y | Z | BSR |
|---|---|---|---|---|---|
| Cerebellum | Right | 6 | −2 | −14.5 | 85.3967 |
| Medial parietal | Right | 4.5 | 10 | −14 | 36.2815 |
| MT | Left | −19.5 | 4.5 | −10.5 | 15.9005 |
| Area 3a/3b | Left | −13 | 16 | 0 | 20.3331 |
| Pulvinar | Left | −6 | 5.5 | 0 | 4.8636 |
| Hippocampus | Right | 13.5 | −4.0 | −4.5 | 10.217 |
| Retrosplenium | Left | −2.0 | 6.5 | −10.0 | 5.567 |
| A1/A2 | Left | −29 | 4.5 | 2 | 5.2371 |
| Area 4 | Left | −4.5 | 21.5 | 4 | 16.7316 |
| Area 6 | Right | 24.5 | 13.5 | 6 | 8.5105 |
| Area 6 | Right | 12.5 | 19.5 | 6.5 | 11.9055 |
| Thalamus | Left | −9.5 | −0.5 | 7.5 | 6.8145 |
| Area 8av | Left | −19.5 | 13.5 | 17.5 | 5.4529 |
| Cerebellum | Right | 11.5 | −11.5 | −21 | −12.8537 |
| Cerebellum | Left | −13.5 | −16.5 | −20.5 | −36.4799 |
| Parietal area PG | Right | 15.5 | 20.5 | 11.5 | −4.8155 |
| POa | Left | −19 | 14.5 | −5.5 | −6.5274 |
| Brainstem | Right | 1 | −20.5 | −3.5 | −12.9475 |
| Pulvinar | Left | −3 | −4.5 | −1.5 | −460.358 |
| TE | Right | 19 | −13.5 | 12 | −22.3974 |
| Thalamus | Left | −2.5 | 3.5 | 9 | −7.623 |
| Hippocampus | Left | −14 | −11.5 | 6.5 | −4.395 |
| Perirhinal | Left | −11 | −17.5 | 6.5 | −7.069 |
| Caudate | Left | −3.5 | 3.5 | 13 | −41.2466 |
| TAa | Left | −21 | −8.5 | 16 | −51.6932 |
| 47 | Right | 20 | 6 | 28 | −5.4324 |
| 10 | Right | 2.5 | 4.5 | 44.5 | −14.8553 |
X, Y, Z, Spatial coordinates in relation to the posterior commissure; BSR, bootstrap ratio values greater than our criterion (BSR > 3, p = 0.005).
Functional connectivity analysis
Four regions were identified as being significantly coactive in our tasks using contrast-task PLS analyses when the monkeys moved through space (CHAIR and WALK conditions): right posterior hippocampus, left caudate nucleus, left orbitofrontal gyrus, and the left posterior cingulate cortex (see Table 4 for coordinates). Movement in large-scale space resulted in a statistically significant pattern (p < 0.05) in functional connectivity pattern from seed-PLS analysis in right posterior hippocampus, left caudate, and the left orbitofrontal gyrus seeds in young monkeys and in the left orbitofrontal gyrus in old monkeys. The left posterior cingulate seed was not statistically significant for either group (p > 0.10). Although it should be pointed out that the strength of the statistically significant relationships may be somewhat inflated due to effects of collinearity, evaluation of the significant seed indicates statistical reliability.
Table 4.
X, Y, Z coordinates for seed locations relative to the posterior commissure
| Area | Hemisphere | X | Y | Z |
|---|---|---|---|---|
| Posterior hippocampus | Right | 14.0 | −5.5 | −3.0 |
| Caudate | Left | −6.5 | 6 | 13.5 |
| Orbitofrontal | Left | −7.5 | 7 | 32.5 |
| Posterior cingulate | Left | −2.0 | 9.5 | −7 |
X, Y, Z, Spatial coordinates in relation to the posterior commissure.
In young monkeys, the right posterior hippocampus seed (Fig. 5) accounted for 54.8% of the covariance (p < 0.001), including significant connectivity with bilateral portions of hippocampus, parahippocampal gyrus, pulvinar, cerebellum, right MST, caudate nucleus, putamen, inferior frontal sulcus, and left cingulate gyrus. In old monkeys, the right posterior hippocampal seed did not show statistically significant (p > 0.05) connectivity with other regions (Fig. 5a). For the young monkey caudate nucleus seed (Fig. 6), 56.5% of the covariance was accounted for (p < 0.01) and included bilateral regions of the hippocampus, anterior cingulate, and several extrahippocampal regions in the temporal lobe. In old monkeys, the caudate nucleus seed did not show significant connectivity with other regions (p > 0.05; Fig. 6a). For the left orbitofrontal cortex, a significant functional connectivity pattern was found, which accounted for 51.69% of the covariance (p < 0.001; Fig. 7) in young and old monkeys. In young monkeys, this region was positively correlated with bilateral posterior hippocampus, caudate nucleus, cerebellum, temporal lobe regions, orbitofrontal regions, and anterior cingulate. The older monkeys showed connectivity with a different set of regions (Fig. 7a), including sensory and motor regions such as the occipital lobe and regions around the central sulcus.
Figure 5.
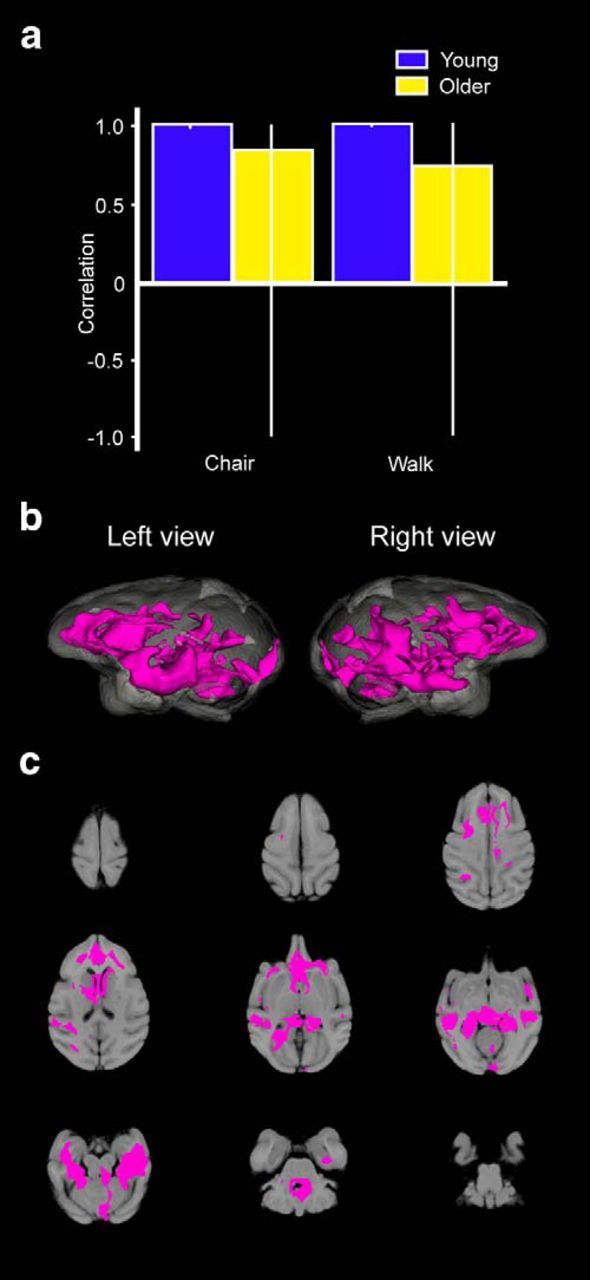
Seed-PLS results for the right posterior hippocampal seed that illustrate regions of the brain that also showed elevated activity during the CHAIR and WALK conditions. Seed-related correlation plots reveal a significant pattern in young monkeys, but not in aged monkeys, that was associated with the right posterior hippocampus (a). The coactivity pattern from the hippocampal seed found in young monkeys is reflected in the magenta regions. No significant pattern was found in older monkeys from this seed. Patterns of FDG metabolism in horizontal slice by slice 2D sections (b) and reconstructed 3D metabolism maps (c) that illustrate the functional connectivity of the posterior hippocampus seed for the young animals. Colors represent values for positive (magenta) and negative (cyan) correlation values (p < 0.005).
Figure 6.

Seed-PLS results for the left caudate nucleus seed that illustrate regions of the brain that also showed elevated activity during the CHAIR and WALK conditions. The coactivity pattern from the caudate seed found in young monkeys is reflected in the magenta regions. No significant pattern was found in older monkeys from this seed. Seed-related correlation plots reveal a significant pattern in young monkeys, but not in aged monkeys (a). Patterns of FDG metabolism in horizontal slice by slice 2D sections (b) and reconstructed 3D metabolism maps (c) that illustrate the functional connectivity of the caudate seed in the young animals.
Figure 7.
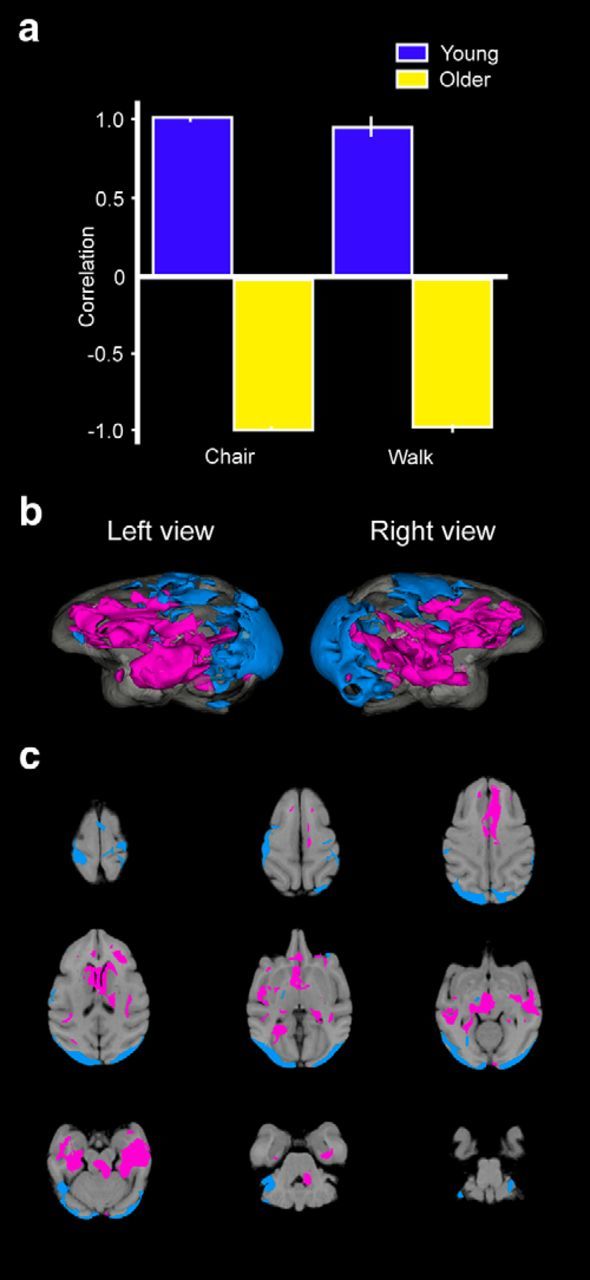
Seed-PLS results for the left orbitofrontal gyrus seed that illustrate regions of the brain that also showed elevated activity during the CHAIR and WALK conditions. Seed-related correlation plots reveal a significant pattern in young monkeys and aged monkeys that was associated the orbitofrontal seed (a). The coactivity pattern from the orbitofrontal seed in young monkeys is reflected in the magenta regions, whereas the coactivity pattern found in older monkeys is reflected in cyan regions. Patterns of FDG metabolism in horizontal slice by slice 2D sections (b) and reconstructed 3D metabolism maps (c) that illustrate the differential functional connectivity of the orbitofrontal seed in young and older animals.
A conjoint, whole-brain connectivity map was created to highlight regions that were coactive across the three significant seed locations in the young monkeys (Fig. 8), which included the right and left posterior hippocampus, left middle hippocampus, regions in the temporal lobe, orbitofrontal, and supplementary sensory areas. During movement in large-scale space, the younger monkeys showed strong connections with many regions across the brain from these seeded regions and much more extensive posterior hippocampal involvement in these behaviors than did the older animals.
Figure 8.
Reconstructed conjoint activity maps illustrate connectivity between the significant hippocampal, caudate, and orbitofrontal seeds in young monkeys. Areas in red highlight the areas that overlapped between the seeds. Conventional coronal sections illustrate specific areas that overlapped between the seeds (a). Reconstructed 3D conjoint maps illustrate the whole-brain connectivity between the seeds (b). White vertical lines indicate the location of the coronal sections found in a.
Potential sources of variance in older monkeys
Task-PLS and seed-PLS analyses consistently found a change in metabolism within the identified networks between young and old monkeys. To explain these differences, we extracted mean metabolism values of the gray matter, cerebellum, and networks identified by LV1 and LV2 for the task PLS. When mean gray matter and cerebellum metabolism values were analyzed by two-way ANOVA, old monkeys had significantly lower mean gray matter metabolism values (F(1,24) = 5.042, p < 0.05; old x = 112.96, SD = 4.48; young x = 117.12, SD = 3.36). No other main effects or interactions were found (p > 0.05). To rule out the possibility that gray matter atrophy was responsible for the measures we used to determine hypometabolism in the older animals, network patterns found in LV1 and LV2 were used to create ROIs from the MRIs (see Materials and Methods) and VBM volumetric analysis was performed. Consistent with other studies comparing young and aged nonhuman primates (Alexander et al., 2008), no significant age difference in gray matter volume was detected in these specific regions (corrected, p > 0.05). Mean metabolism values were also extracted from these regions and three-way ANOVA found significant main effects for age-related hypometabolism in the networks identified by LV1 (F(1,48) = 6.81, p < 0.05) and LV2 (F(1,48) = 311.99, p < 0.05), confirming the PLS results above. No other main effects or interactions were found (p > 0.05).
Discussion
Using [18F]FDG-microPET and PLS correlation analysis, we found, within individual young and old rhesus macaques, distinct patterns of brain metabolic activity in response to four levels of engagement in navigation behaviors. Two primary novel findings emerge from this experiment. First we identify whole-brain activity patterns in the nonhuman primate that are specifically associated with constrained (CAGE and TREADMILL), restrained (CHAIR), and unconstrained (WALK) locomotion. Second, we demonstrate that aging alters the metabolic participation of networks activated by these behaviors and, in some cases, their composition.
It is well known that locomotion through space modulates hippocampal principal cell activity powerfully in rats (O'Keefe and Dostrovsky, 1971) and that the stability of hippocampal cognitive map representations is altered in aging (Barnes et al., 1997; Wilson et al., 2005). In fact, the expression of normal place-cell-receptive fields are altered dramatically in rat hippocampus when movement is restrained (Foster et al., 1989), although the ability to locomote on a ball can mitigate these changes despite head restraint (Chen et al., 2013). In addition, when rats are moved passively through space on robots (Gavrilov et al., 1998) within the confines of a cart (Winter et al., 2015b) or are trained to “drive” a car on a track (Terrazas et al., 2005), hippocampal and entorhinal cell firing patterns are clearly degraded. Optic flow (Gavrilov et al., 1998; Terrazas et al., 2005) and vestibular (Stackman et al., 2002) signals are also known to contribute to the temporal lobe networks involved in locomotion through space. As discussed below, the present data suggest that there may be an impact on activity levels of recruited neural circuits or altered participation of networks when locomotion or other sensory signals are restricted. This may have implications for neural activity measured in scanner environments for humans or under head-fixed restraint conditions during electrophysiological recording in nonhuman primates.
Although there have been a number of studies examining brain activity in relation to gait, there have only been two functional imaging studies that have examined locomotion through space in an unconstrained manner similar to the present study. For example, one PET study examined brain activity after running (Tashiro et al., 2001) and found increased activity levels in the occipital lobe, posterior parietal lobe, precentral gyrus, postcentral gyrus, and the cerebellum; however, only selected brain regions were measured and the design did not use a nonrunning control condition. The other study used PET to scan participants after brief periods of walking and fMRI to scan during imagined locomotion (la Fougère et al., 2010). Both real and imagined locomotion engaged regions of frontal and occipital cortex, parahippocampal gyri, and the cerebellum, but were distinct in that real locomotion was associated with increased activity in primary motor and sensory areas, whereas imagined locomotion was associated with increased activity in supplemental motor areas and regions of the basal ganglia. The unconstrained locomotion condition in the present study activated regions consistent with these human studies with the addition of MT, MST, retrosplenial cortex, posterior cingulate cortex, right posterior hippocampus, right colliculi, left pulvinar, and the thalamus. Detection of these additional areas is attributable to our experimental design and the use of PLS correlation analysis to detect coactivity relationships across the brain. We also intensity normalized our data relative to white matter FDG uptake rather than with respect to global mean uptake, which facilitates detection of small changes in FDG metabolism (Borghammer et al., 2008).
One strength of the present study was the ability to compare unconstrained locomotion in space (WALK) with both constrained locomotion (CAGE, TREADMILL) and restrained passive locomotion through space (CHAIR). Activity patterns generated by these distinct movement conditions involved, to differing degrees, regions of the hippocampus, striatum, cerebellum, and occipital, parietal, temporal, and frontal lobes. We know that, in humans, mental imagery of walking and running can activate bilateral hippocampus and that the right parahippocampal and fusiform gyri are involved in visually guided locomotion (Jahn et al., 2008, 2009). We also know that the parahippocampus is required for spatially guided behaviors and spatial learning in humans (Aguirre et al., 1996; Bohbot and Corkin, 2007) and that selective inactivation of components of navigation networks in rats disrupt grid cell activity in the parahippocampus (Winter et al., 2015a). In the present study, we found that the parahippocampus was active in those conditions that involved movement through space (i.e., WALK and CHAIR), but was not active during the conditions that involved locomotion (i.e., WALK and TREADMILL). These data support observations that the parahippocampus is involved in visually guided movement through space and suggest that it is not involved in motor-driven locomotion.
Although the brain regions coactivated during the four behavioral conditions were similar across age, the network activity patterns in older macaques did not dissociate robustly between these distinct behaviors. That is, under the present behavioral conditions, the age difference observed was not related to more brain regions being recruited to facilitate behavior, but rather to a decrease in the differentiation between activity in each network that was associated with the different movement conditions. The present data suggesting less specialized networks for spatial navigation are reminiscent of observations made using MRI methods in object recognition pathways of older adults (Park et al., 2004; Ryan et al., 2012). The present data are also consistent with human aging studies suggesting that that networks can become effectively “dedifferentiated” by losing their activation specificity for select behaviors or cognitive processes (Goh, 2011) and with fMRI studies showing reduced activity across similar regions during virtual navigation in aged humans (Moffat et al., 2006; Antonova et al., 2009). Although the right posterior hippocampus was engaged in large-scale spatial movement in both age groups, this structure was more weakly active in the older macaques. The activity observed in the hippocampus in both age groups in the present study is anatomically consistent with human lesion (Abrahams et al., 1997; Bohbot et al., 1998) and electrophysiological and MRI data collected during virtual navigation (Burgess et al., 2002; Jacobs et al., 2010; Kaplan et al., 2012).
The present experiment identified circuits that were distinctly coactive in the CHAIR or WALK conditions, which separated a network that was optic flow specific (i.e., CHAIR condition) from a network that was self-motion specific (i.e., WALK condition). Four of the areas coactivated in the WALK and CHAIR conditions were targeted for functional connectivity analysis. The posterior cingulate cortex was of interest because of its association with the default mode network (DMN) system (Greicius et al., 2003; Fox et al., 2005; Buckner and Vincent, 2007; Vincent et al., 2007), which shows reduced connectivity in aged adults (Tomasi and Volkow, 2012; Ferreira and Busatto, 2013; Vidal-Piñeiro et al., 2014). The locomotion behaviors examined in the present study, however, did not reveal significant connections across the brain from this seed in either age group. Disconnection during locomotion may be analogous to temporal lobe structures becoming decoupled from the DMN during episodic encoding (Buckner et al., 2008).
There were significant connections revealed from the orbitofrontal seed in both age groups. Although the younger animals showed connections with limbic areas from the orbitofrontal seed, the correlated patterns of activity were with sensory and supplemental sensory areas in the aged macaques. Whether the different functional connections in the old compared with young monkeys in the orbitofrontal seed analysis reflect recruitment of additional circuits to ensure adaptive behavioral outcomes or dysfunctional recruitment of circuits not optimal for performance remains an open question. The posterior shift found in the orbitofrontal seed, however, is at least consistent with the idea that the older macaques may use sensory-based strategies to guide their behavior during movement in space (Davis et al., 2008).
When the right posterior hippocampus was the targeted seed, only the young monkeys showed significant functional connections to widespread areas of the temporal lobe and ventral regions of the occipital and frontal lobes. For the young monkeys, the left caudate nucleus and the left orbitofrontal gyrus displayed similar patterns of functional connections. Therefore, a conjoint connectivity map was constructed across these regions for the young animals (Fig. 8) and shows a core network including right and left posterior hippocampus, left middle hippocampus, large regions in the temporal lobe, orbitofrontal, and several supplementary sensory areas. It is possible that the small sample size within groups generated collinearity effects and inflated the functional connectivity patterns for the three significant seeds in the younger macaques and the one significant seed in the older macaques. Future studies with larger within-group samples are necessary to confirm the observed functional connectivity patterns and to determine whether additional connected areas can be detected.
Overall, the lack of well differentiated behavioral networks, reduced metabolic values in the active areas, and corresponding changes in functional connectivity may contribute to the reported age-related deficits in spatial cognition. Future studies with larger group sizes and that manipulate network activity and functional connections directly are needed to confirm this conclusion.
Footnotes
This work was supported by the National Institutes of Health (Grants R01 AG003376 and P51 RR000169) and the McKnight Brain Research Foundation. We thank the staff of the Center for Molecular and Genomic Imaging, University of California–Davis, for assistance with imaging studies and Drs. Kewei Chen, Eric Reiman, and Arne Ekstrom for helpful comments on the manuscript.
The authors declare no competing financial interests.
References
- Abrahams S, Pickering A, Polkey CE, Morris RG. Spatial memory deficits in patients with unilateral damage to the right hippocampal formation. Neuropsychologia. 1997;35:11–24. doi: 10.1016/S0028-3932(96)00051-6. [DOI] [PubMed] [Google Scholar]
- Aguirre GK, Detre JA, Alsop DC, D'Esposito M. The parahippocampus subserves topographical learning in man. Cereb Cortex. 1996;6:823–829. doi: 10.1093/cercor/6.6.823. [DOI] [PubMed] [Google Scholar]
- Alexander GE, Chen K, Aschenbrenner M, Merkley TL, Santerre-Lemmon LE, Shamy JL, Skaggs WE, Buonocore MH, Rapp PR, Barnes CA. Age-related regional network of magnetic resonance imaging gray matter in the rhesus macaque. J Neurosci. 2008;28:2710–2718. doi: 10.1523/JNEUROSCI.1852-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonova E, Parslow D, Brammer M, Dawson GR, Jackson SH, Morris RG. Age-related neural activity during allocentric spatial memory. Memory. 2009;17:125–143. doi: 10.1080/09658210802077348. [DOI] [PubMed] [Google Scholar]
- Barnes CA. Memory deficits associated with senescence: a neurophysiological and behavioral study in the rat. J Comp Physiol Psychol. 1979;93:74–104. doi: 10.1037/h0077579. [DOI] [PubMed] [Google Scholar]
- Barnes CA, Suster MS, Shen J, McNaughton BL. Multistability of cognitive maps in the hippocampus of old rats. Nature. 1997;388:272–275. doi: 10.1038/40859. [DOI] [PubMed] [Google Scholar]
- Bohbot VD, Corkin S. Posterior parahippocampal place learning in H.M. Hippocampus. 2007;17:863–872. doi: 10.1002/hipo.20313. [DOI] [PubMed] [Google Scholar]
- Bohbot VD, Kalina M, Stepankova K, Spackova N, Petrides M, Nadel L. Spatial memory deficits in patients with lesions to the right hippocampus and to the right parahippocampal cortex. Neuropsychologia. 1998;36:1217–1238. doi: 10.1016/S0028-3932(97)00161-9. [DOI] [PubMed] [Google Scholar]
- Bohbot VD, Lerch J, Thorndycraft B, Iaria G, Zijdenbos AP. Gray matter differences correlate with spontaneous strategies in a human virtual navigation task. J Neurosci. 2007;27:10078–10083. doi: 10.1523/JNEUROSCI.1763-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghammer P, Jonsdottir KY, Cumming P, Ostergaard K, Vang K, Ashkanian M, Vafaee M, Iversen P, Gjedde A. Normalization in PET group comparison studies-the importance of a valid reference region. Neuroimage. 2008;40:529–540. doi: 10.1016/j.neuroimage.2007.12.057. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Vincent JL. Unrest at rest: Default activity and spontaneous network correlations. Neuroimage. 2007;37:1091–1096. doi: 10.1016/j.neuroimage.2007.01.010. discussion 1097–1099. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Andrews-Hanna JR, Schacter DL. The brain's default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- Burgess N, Maguire EA, O'Keefe J. The human hippocampus and spatial and episodic memory. Neuron. 2002;35:625–641. doi: 10.1016/S0896-6273(02)00830-9. [DOI] [PubMed] [Google Scholar]
- Cabeza R, McIntosh AR, Tulving E, Nyberg L, Grady CL. Age-related differences in effective neural connectivity during encoding and recall. Neuroreport. 1997;8:3479–3483. doi: 10.1097/00001756-199711100-00013. [DOI] [PubMed] [Google Scholar]
- Chen G, King JA, Burgess N, O'Keefe J. How vision and movement combine in the hippocampal place code. Proc Natl Acad Sci U S A. 2013;110:378–383. doi: 10.1073/pnas.1215834110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SW, Dennis NA, Daselaar SM, Fleck MS, Cabeza R. Qué PASA? The posterior-anterior shift in aging. Cereb Cortex. 2008;18:1201–1209. doi: 10.1093/cercor/bhm155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrom A, Suthana N, Millett D, Fried I, Bookheimer S. Correlation between BOLD fMRI and theta-band local field potentials in the human hippocampal area. J Neurophysiol. 2009;101:2668–2678. doi: 10.1152/jn.91252.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov A, Li X, Pohl KM, Bouix S, Styner M, Addicott M, Wyatt C, Daunais JB, Wells WM, Kikinis R. Atlas-guided segmentation of vervet monkey brain MRI. Open Neuroimaging J. 2011;5:186–197. doi: 10.2174/1874440001105010186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira LK, Busatto GF. Resting-state functional connectivity in normal brain aging. Neurosci Biobehav Rev. 2013;37:384–400. doi: 10.1016/j.neubiorev.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Foster TC, Castro CA, McNaughton BL. Spatial selectivity of rat hippocampal neurons: dependence on preparedness for movement. Science. 1989;244:1580–1582. doi: 10.1126/science.2740902. [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilov VV, Wiener SI, Berthoz A. Discharge correlates of hippocampal complex spike neurons in behaving rats passively displaced on a mobile robot. Hippocampus. 1998;8:475–490. doi: 10.1002/(SICI)1098-1063(1998)8:5<475::AID-HIPO7>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Goense J, Logothetis NK, Merkle H. Flexible, phase-matched, linear receive arrays for high-field MRI in monkeys. Magn Reson Imaging. 2010;28:1183–1191. doi: 10.1016/j.mri.2010.03.026. [DOI] [PubMed] [Google Scholar]
- Goh JO. Functional dedifferentiation and altered connectivity in older adults: neural accounts of cognitive aging. Aging Dis. 2011;2:30–48. [PMC free article] [PubMed] [Google Scholar]
- Grady CL, Grigg O, Ng C. Age differences in default and reward networks during processing of personally relevant information. Neuropsychologia. 2012;50:1682–1697. doi: 10.1016/j.neuropsychologia.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci U S A. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley T, Maguire EA, Spiers HJ, Burgess N. The well-worn route and the path less traveled: distinct neural bases of route following and wayfinding in humans. Neuron. 2003;37:877–888. doi: 10.1016/S0896-6273(03)00095-3. [DOI] [PubMed] [Google Scholar]
- Head D, Isom M. Age effects on wayfinding and route learning skills. Behav Brain Res. 2010;209:49–58. doi: 10.1016/j.bbr.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Holschneider DP, Maarek JM. Brain maps on the go: functional imaging during motor challenge in animals. Methods. 2008;45:255–261. doi: 10.1016/j.ymeth.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J, Kahana MJ, Ekstrom AD, Mollison MV, Fried I. A sense of direction in human entorhinal cortex. Proc Natl Acad Sci U S A. 2010;107:6487–6492. doi: 10.1073/pnas.0911213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn K, Deutschländer A, Stephan T, Kalla R, Wiesmann M, Strupp M, Brandt T. Imaging human supraspinal locomotor centers in brainstem and cerebellum. Neuroimage. 2008;39:786–792. doi: 10.1016/j.neuroimage.2007.09.047. [DOI] [PubMed] [Google Scholar]
- Jahn K, Wagner J, Deutschländer A, Kalla R, Hüfner K, Stephan T, Strupp M, Brandt T. Human hippocampal activation during stance and locomotion: fMRI study on healthy, blind, and vestibular-loss subjects. Ann N Y Acad Sci. 2009;1164:229–235. doi: 10.1111/j.1749-6632.2009.03770.x. [DOI] [PubMed] [Google Scholar]
- Kaplan R, Doeller CF, Barnes GR, Litvak V, Düzel E, Bandettini PA, Burgess N. Movement-related theta rhythm in humans: Coordinating self-directed hippocampal learning. PLoS Biol. 2012;10:e1001267. doi: 10.1371/journal.pbio.1001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi K, Bohbot VD. Spatial navigational strategies correlate with gray matter in the hippocampus of healthy older adults tested in a virtual maze. Front Aging Neurosci. 2013;5:1. doi: 10.3389/fnagi.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A, Williams LJ, McIntosh AR, Abdi H. Partial Least Squares (PLS) methods for neuroimaging: a tutorial and review. Neuroimage. 2011;56:455–475. doi: 10.1016/j.neuroimage.2010.07.034. [DOI] [PubMed] [Google Scholar]
- la Fougère C, Zwergal A, Rominger A, Förster S, Fesl G, Dieterich M, Brandt T, Strupp M, Bartenstein P, Jahn K. Real versus imagined locomotion: a [18F]-FDG PET-fMRI comparison. Neuroimage. 2010;50:1589–1598. doi: 10.1016/j.neuroimage.2009.12.060. [DOI] [PubMed] [Google Scholar]
- Maguire EA, Gadian DG, Johnsrude IS, Good CD, Ashburner J, Frackowiak RS, Frith CD. Navigation-related structural change in the hippocampi of taxi drivers. Proc Natl Acad Sci U S A. 2000;97:4398–4403. doi: 10.1073/pnas.070039597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh AR, Lobaugh NJ. Partial least squares analysis of neuroimaging data: applications and advances. Neuroimage. 2004;23:S250–263. doi: 10.1016/j.neuroimage.2004.07.020. [DOI] [PubMed] [Google Scholar]
- McIntosh AR, Bookstein FL, Haxby JV, Grady CL. Spatial pattern analysis of functional brain images using partial least squares. Neuroimage. 1996;3:143–157. doi: 10.1006/nimg.1996.0016. [DOI] [PubMed] [Google Scholar]
- McIntosh AR, Sekuler AB, Penpeci C, Rajah MN, Grady CL, Sekuler R, Bennett PJ. Recruitment of unique neural systems to support visual memory in normal aging. Curr Biol. 1999;9:1275–1278. doi: 10.1016/S0960-9822(99)80512-0. [DOI] [PubMed] [Google Scholar]
- Moffat SD, Resnick SM. Effects of age on virtual environment place navigation and allocentric cognitive mapping. Behav Neurosci. 2002;116:851–859. doi: 10.1037/0735-7044.116.5.851. [DOI] [PubMed] [Google Scholar]
- Moffat SD, Zonderman AB, Resnick SM. Age differences in spatial memory in a virtual environment navigation task. Neurobiol Aging. 2001;22:787–796. doi: 10.1016/S0197-4580(01)00251-2. [DOI] [PubMed] [Google Scholar]
- Moffat SD, Elkins W, Resnick SM. Age differences in the neural systems supporting human allocentric spatial navigation. Neurobiol Aging. 2006;27:965–972. doi: 10.1016/j.neurobiolaging.2005.05.011. [DOI] [PubMed] [Google Scholar]
- O'Keefe J, Dostrovsky J. The hippocampus as a spatial map: preliminary evidence from unit activity in the freely-moving rat. Brain Res. 1971;34:171–175. doi: 10.1016/0006-8993(71)90358-1. [DOI] [PubMed] [Google Scholar]
- Park DC, Polk TA, Park R, Minear M, Savage A, Smith MR. Aging reduces neural specialization in ventral visual cortex. Proc Natl Acad Sci U S A. 2004;101:13091–13095. doi: 10.1073/pnas.0405148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl KM, Bouix S, Nakamura M, Rohlfing T, McCarley RW, Kikinis R, Grimson WE, Shenton ME, Wells WM. A hierarchical algorithm for MR brain image parcellation. IEEE Trans Med Imaging. 2007;26:1201–1212. doi: 10.1109/TMI.2007.901433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser BA, Koopmans PJ, Witzel T, Wald LL, Barth M. Three dimensional echo-planar imaging at 7 Tesla. Neuroimage. 2010;51:261–266. doi: 10.1016/j.neuroimage.2010.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp PR, Kansky MT, Roberts JA. Impaired spatial information processing in aged monkeys with preserved recognition memory. Neuroreport. 1997;8:1923–1928. doi: 10.1097/00001756-199705260-00026. [DOI] [PubMed] [Google Scholar]
- Rosenzweig ES, Redish AD, McNaughton BL, Barnes CA. Hippocampal map realignment and spatial learning. Nat Neurosci. 2003;6:609–615. doi: 10.1038/nn1053. [DOI] [PubMed] [Google Scholar]
- Ryan L, Cardoza JA, Barense MD, Kawa KH, Wallentin-Flores J, Arnold WT, Alexander GE. Age-related impairment in a complex object discrimination task that engages perirhinal cortex. Hippocampus. 2012;22:1978–1989. doi: 10.1002/hipo.22069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RD, Barnes CA. Impact of aging brain circuits on cognition. Eur J Neurosci. 2013;37:1903–1915. doi: 10.1111/ejn.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaggs WE, McNaughton BL, Permenter M, Archibeque M, Vogt J, Amaral DG, Barnes CA. EEG sharp waves and sparse ensemble unit activity in the macaque hippocampus. J Neurophysiol. 2007;98:898–910. doi: 10.1152/jn.00401.2007. [DOI] [PubMed] [Google Scholar]
- Stackman RW, Clark AS, Taube JS. Hippocampal spatial representations require vestibular input. Hippocampus. 2002;12:291–303. doi: 10.1002/hipo.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro M, Itoh M, Fujimoto T, Fujiwara T, Ota H, Kubota K, Higuchi M, Okamura N, Ishii K, Bereczki D, Sasaki H. 18F-FDG PET mapping of regional brain activity in runners. J Sports Med Phys Fitness. 2001;41:11–17. [PubMed] [Google Scholar]
- Terrazas A, Krause M, Lipa P, Gothard KM, Barnes CA, McNaughton BL. Self-motion and the hippocampal spatial metric. J Neurosci. 2005;25:8085–8096. doi: 10.1523/JNEUROSCI.0693-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND. Aging and functional brain networks. Mol Psychiatry. 2012;17:471, 549–558. doi: 10.1038/mp.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Piñeiro D, Valls-Pedret C, Fernández-Cabello S, Arenaza-Urquijo EM, Sala-Llonch R, Solana E, Bargalló N, Junqué C, Ros E, Bartrés-Faz D. Decreased default mode network connectivity correlates with age-associated structural and cognitive changes. Front Aging Neurosci. 2014;6:256. doi: 10.3389/fnagi.2014.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Wiener JM, de Condappa O, Harris MA, Wolbers T. Maladaptive bias for extrahippocampal navigation strategies in aging humans. J Neurosci. 2013;33:6012–6017. doi: 10.1523/JNEUROSCI.0717-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IA, Ikonen S, Gallagher M, Eichenbaum H, Tanila H. Age-associated alterations of hippocampal place cells are subregion specific. J Neurosci. 2005;25:6877–6886. doi: 10.1523/JNEUROSCI.1744-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, McNaughton BL. Dynamics of the hippocampal ensemble code for space. Science. 1993;261:1055–1058. doi: 10.1126/science.8351520. [DOI] [PubMed] [Google Scholar]
- Winter SS, Clark BJ, Taube JS. Disruption of the head direction cell network impairs the parahippocampal grid cell signal. Science. 2015a;347:870–874. doi: 10.1126/science.1259591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SS, Mehlman ML, Clark BJ, Taube JS. Passive transport disrupts grid signals in the parahippocampal cortex. Curr Biol. 2015b;25:2493–2502. doi: 10.1016/j.cub.2015.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisco JJ, Killiany RJ, Guttmann CR, Warfield SK, Moss MB, Rosene DL. An MRI study of age-related white and gray matter volume changes in the rhesus monkey. Neurobiol Aging. 2008;29:1563–1575. doi: 10.1016/j.neurobiolaging.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]