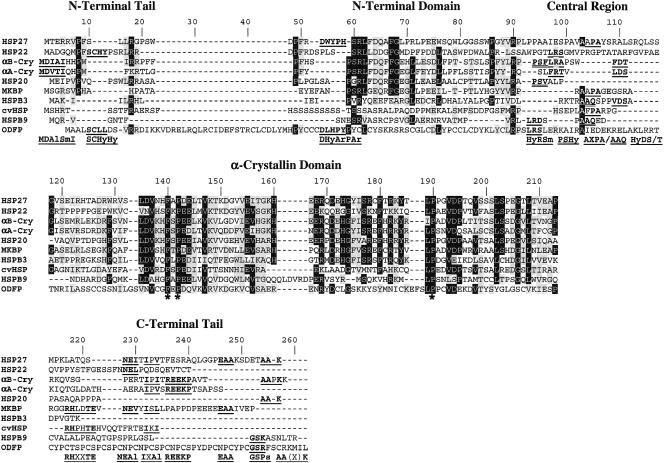
Fig. 1.
Multiple alignment of the known 10 human small heat shock proteins. The alignment was done using CLUSTALW and improved by manual editing. Identical amino acid residues are highlighted in black or gray if they occur in at least 6 or 5 of the 10 sequences, respectively. Similar amino acid residues (E/D, A/G, H/F/W/Y, S/T, I/L/V, H/R/K) are highlighted in gray if they occur in at least 1 of the remaining sequences or in at least 5 of the 10 sequences. Sequence motifs in the variable regions common to at least 2 sequences are in bold and underlined. The corresponding consensus motifs are given in the bottom line using the amino acid groupings as described in the Materials and Methods.