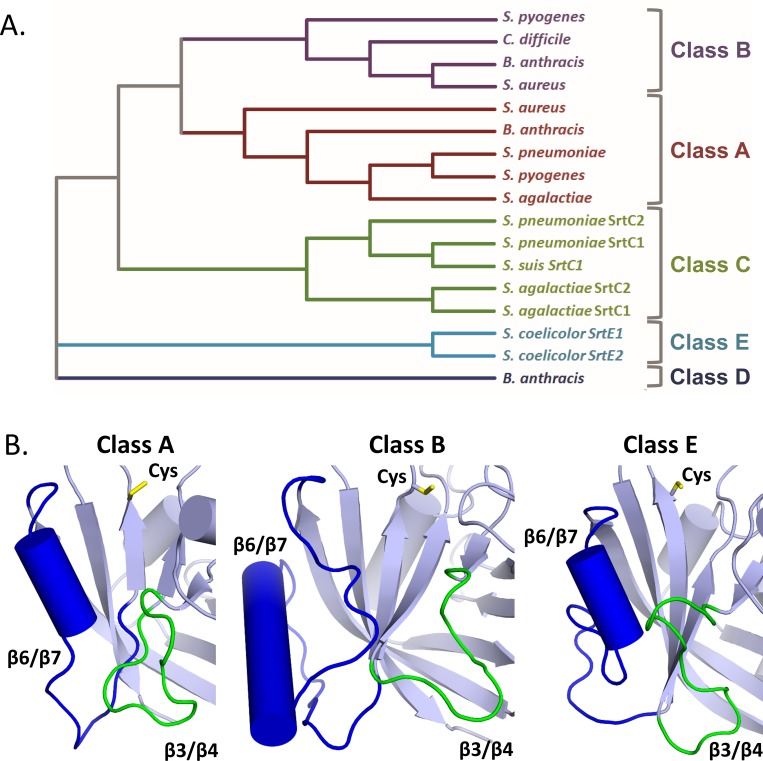
Fig 3. The structure of SrtE1 reveals unique features within Class E sortases.
A) Phylogenetic tree of distinct sortase classes. The full amino acid sequences of 17 structurally characterized sortase enzymes were aligned with the sequences of SrtE1 and SrtE2 using the MUSCLE server and submitted to the ClustalOmega program for phylogenetic tree generation via the neighbor joining method [43,45]. The bacterial species and accession numbers of the amino acid sequences used are as follows: Streptomyces coelicolor (NP_628038; NP_628037), Bacillus anthracis (WP_011732503; WP_000093563; WP_000771607), Streptococcus pyogenes (WP_002984641; WP_010921812), Clostridioides difficile (WP_021376017), Staphylococcus aureus (WP_000759361; WP_054104750), Streptococcus agalactiae (WP_017646311; WP_000529911; WP_000746226), Streptococcus pneumoniae (WP_000078846; WP_001140539; ABS82110), and Streptococcus suis (WP_012027975). B) Sortase class comparison of distinguishing class E structural features. Class A and class B enzymes from S. aureus and a class E enzyme (SrtE1) from S. coelicolor are shown in cartoon with helices (cylinders), β3/β4 loops (green), β6/β7 loops (blue), and active site cysteine residue (yellow) indicated. Inspection of the most structurally related class C (PDB ID: 3RE9) and class D (PDB ID: 2LN7) enzymes, as determined by DALI analysis, revealed β3/β4 and β6/β7 loops that align similarly to the class A enzyme.