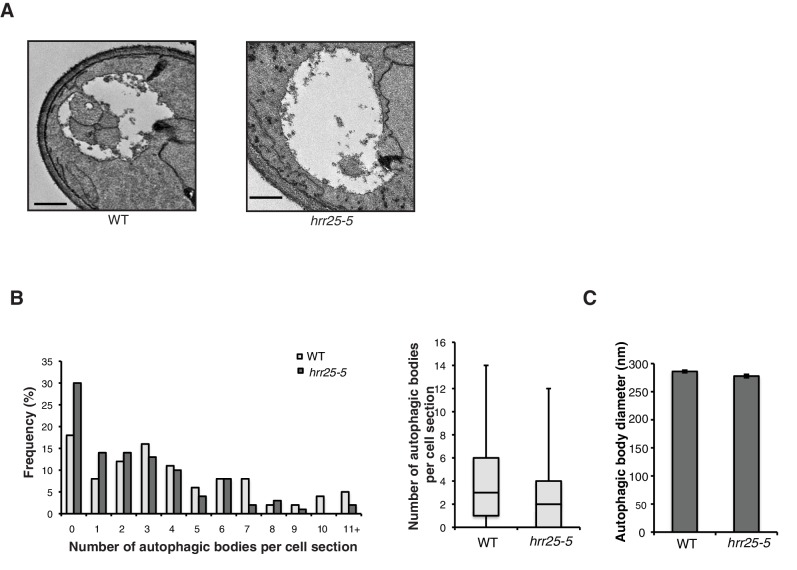
Figure 5. Phosphorylation of the Sec24 membrane distal sites regulates the Sec24-Atg9 interaction.
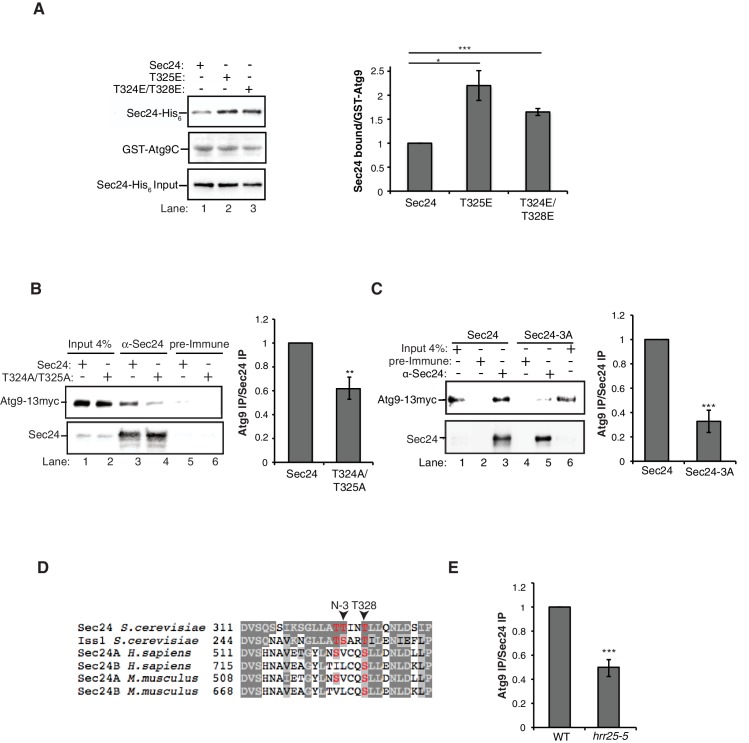
(A) GST-Atg9C (200 nM) was incubated with 37.5 nM of WT Sec23/Sec24-His6, Sec23/Sec24-T325E-His6 or Sec23/Sec24-T324E/T328E-His6 (left). Ratio of Sec24 bound to GST-Atg9C was quantified from three biological replicates. Averages and s.e.m. are shown (right). WT Sec23/Sec24 was set as one for each experiment; p-value = 0.018 (T325E), 0.0008 (T324E/T328E), Student’s unpaired t-test. (B) Sec24 (WT) and Sec24-T324A/T325A or (C) Sec24-T324A/T325A/T328A (Sec24-3A) were immunoprecipitated from lysates expressing Atg9-13myc as described in the Materials and Methods. Precipitated Atg9-13myc was quantitated and normalized to the amount of Sec24 in the precipitate. WT Sec24 was set as one for each experiment. Averages and s.e.m. are shown for 4 (B) or 5 (C) biological replicates; p-value = 0.006 (B), 0.0008 (C) Student’s unpaired t-test. (D) Alignment of the region surrounding T324/T325/T328 (shown in red) with Sec24 orthologues. (E) Same as (B) except Sec24 was immunoprecipitated from WT or hrr25-5 lysates. Averages and s.e.m. are shown for five biological replicates. p-value = 0.0002, Student’s unpaired t-test. **p<0.01; ***p<0.01; ***p<0.001.
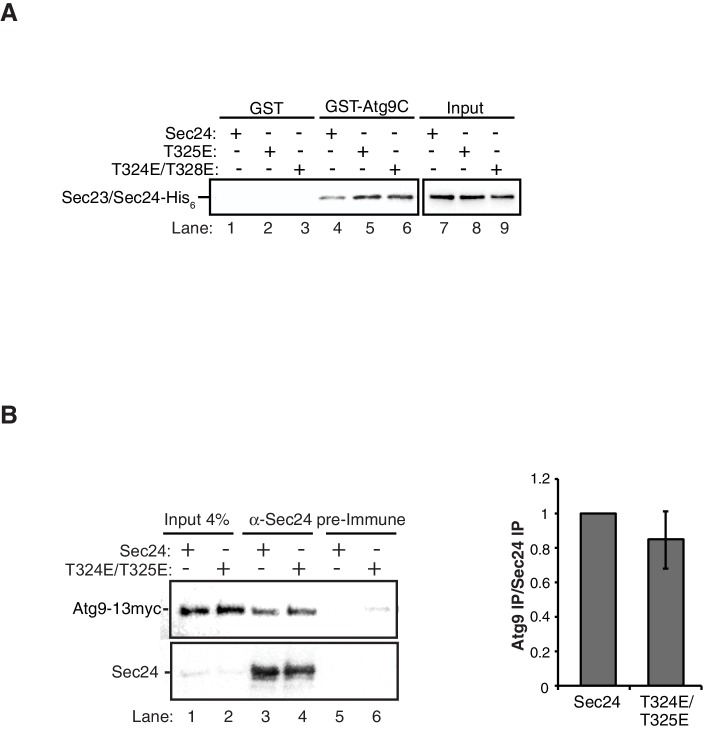
Figure 5—figure supplement 1. GST negative control for in vitro binding in Figure 5A.
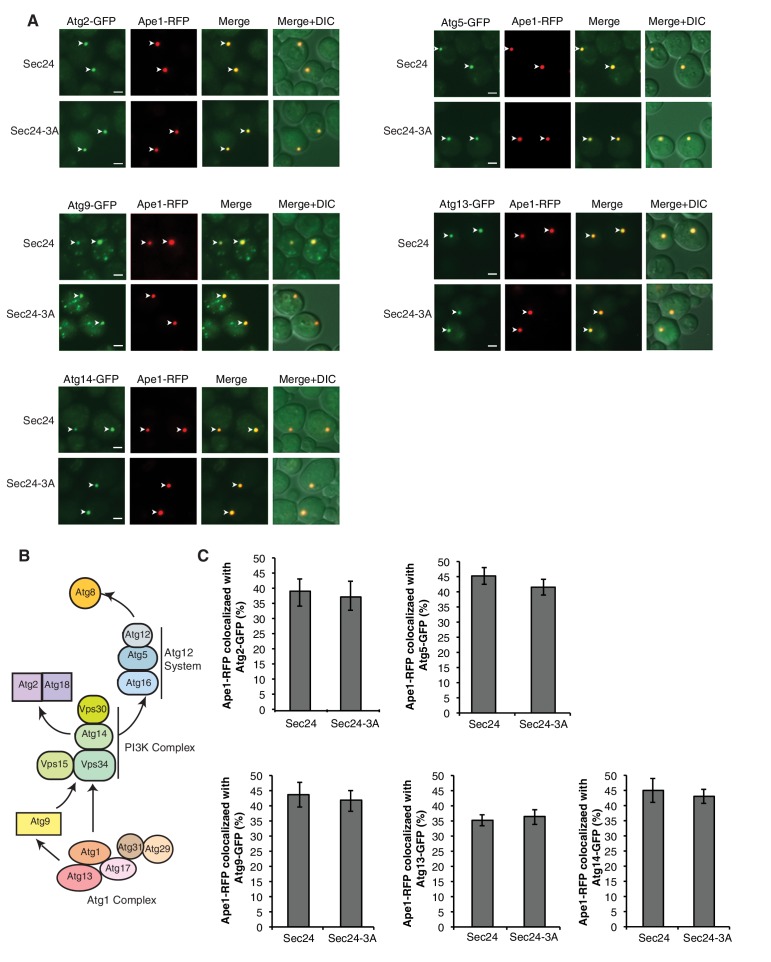
Figure 5—figure supplement 2. Sec24-3A does not affect Atg assembly at the PAS.
Figure 5—figure supplement 3. Sec24-3A does not affect ERES formation.
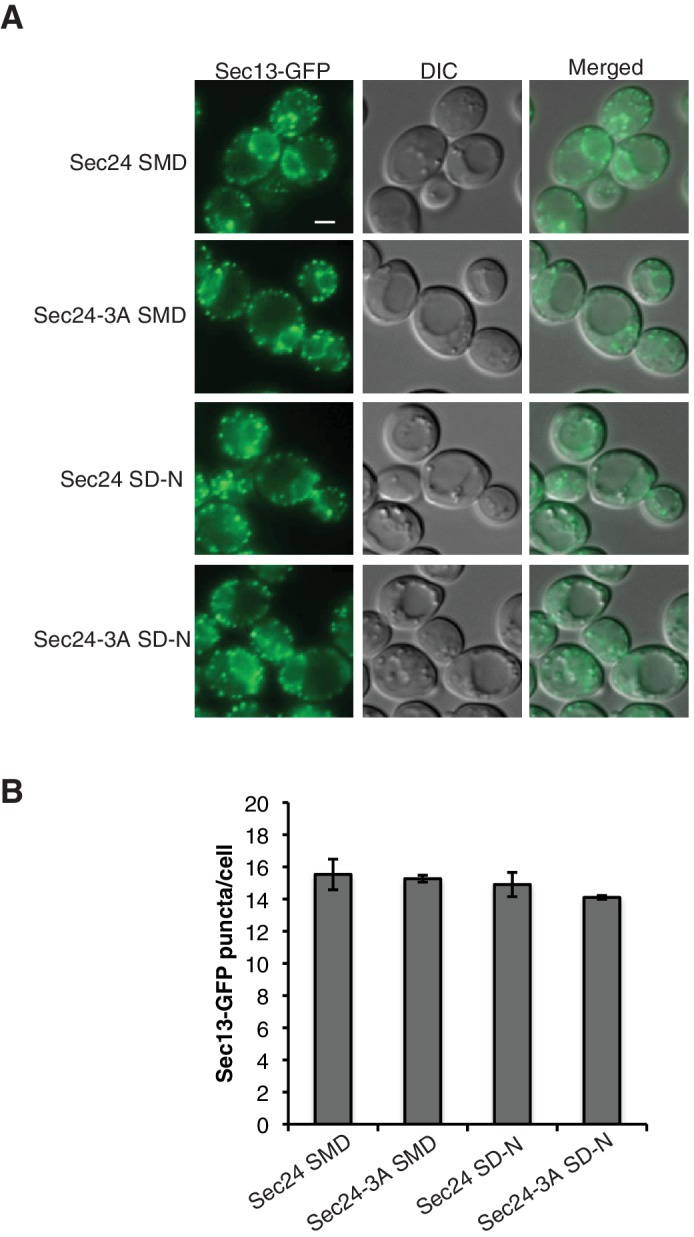
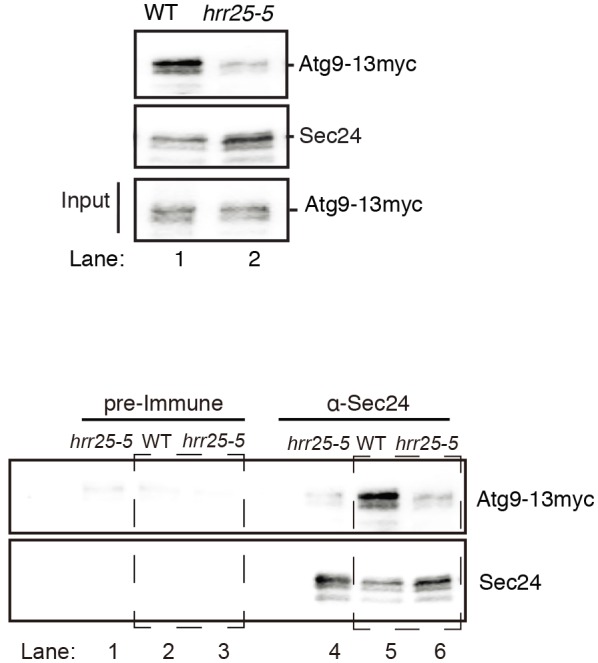
Figure 5—figure supplement 4. Representative blots for quantitation shown in Figure 5E.