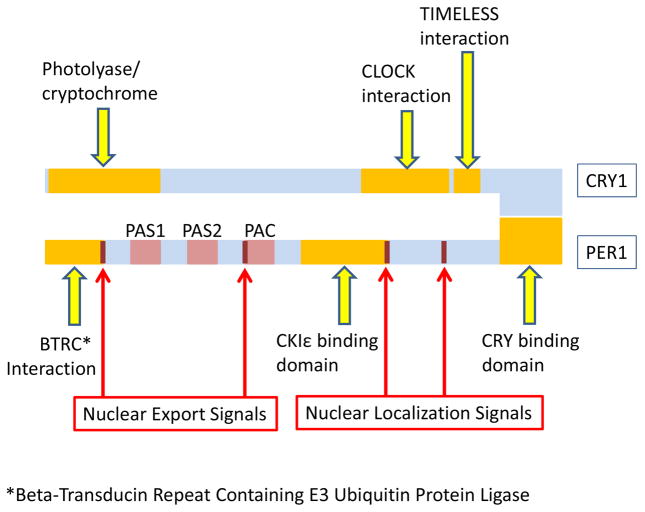
Figure 2. General structure of core clock genes Per and Cry.
The three Per genes encode proteins that contain the PER–ARNT–SIM (PAS) and the related PAC motifs and form oligomers with the Cry proteins to form functional DNA binding complexes. Per1 contains a BTRC interaction domain at the N terminal end. In addition, Per proteins also contain a phosphorylation domain where Casein kinase 1 binds and phosphorylates these proteins. The very C terminus contains a specific domain that allows oligomerization with the Cry proteins. In addition, Per2 contains a PPAR gamma binding site between the CKIε and CRY binding sites that is not seen in Per1 or Per3. Per2 also has a stabilization domain upstream of the PAS motifs not seen in other Per proteins. All Per proteins, however, express nuclear localization and export sequences that allows them to enter and leave the nucleus as needed. The structure of the Cry proteins is much less complex. Both Cry1 and Cry2 contain a cryptochrome (flavoproteins that are sensitive to blue light) interaction domain, as well as a domain that allow them to interact with and inhibit the CLOCK/BMAL1 dimers. In addition, Cry1 also contain a domain that allows interaction with TIMELESS (which negatively regulates CLOCK/BMAL-1 interactions), but this domain is not found in Cry2. (Based on http://www.uniprot.org/).