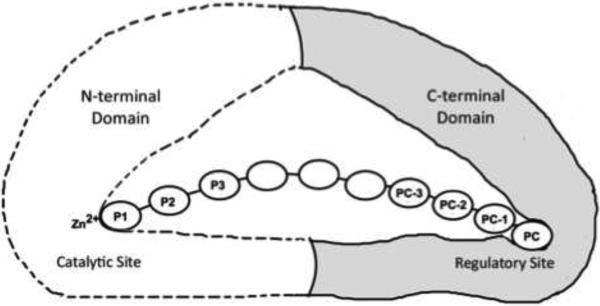
Figure 1. Allosteric activation of ERAP1.
ERAP1 trims peptide precursors by a molecular ruler mechanism. To form a productive complex, the peptide substrate makes a concurrent binding of its N- and C-termini to the catalytic and the regulatory sites of ERAP1, respectively. This allosteric binding changes ERAP1 into a high-activity closed conformation, and efficiently trims one residue from the peptide's N-terminus. The peptide substrate's residue positions are numbered forward from the N-terminus as P1, P2, P3, and backward from the C-terminus as peptide carboxyl-terminal PC, the penultimate PC-1, and further towards the N-terminal direction PC-2, PC-3 etc.