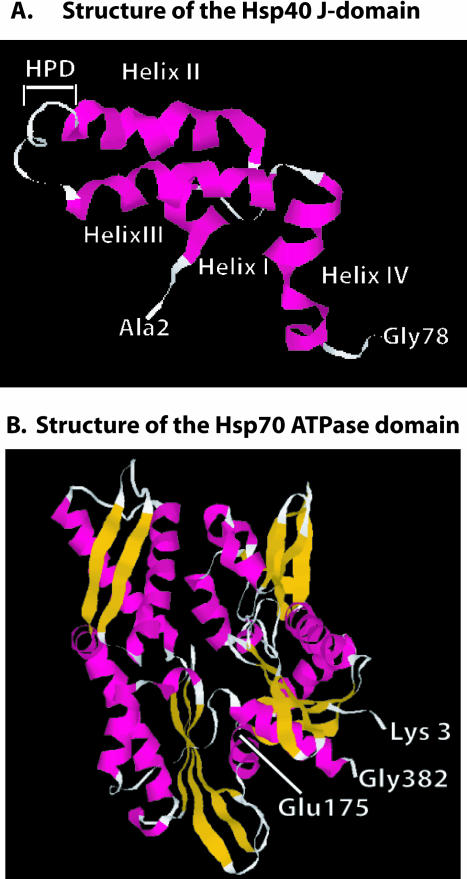
Fig 3.
Structures of the Hsp40 J-domain and the Hsp70 adenosine triphosphatase (ATPase) domain. (A) Ribbon diagram of the nuclear magnetic resonance solution structure of residues 2–77 of E coli DnaJ. HPD denotes the position of the conserved HPD motif that is found in the J-domain of all Hsp40s. D in the HPD motif corresponds to D35 in DnaJ that was mutated in experiments that identified the J-domain–binding site in DnaK (see text for details). Gly78 denotes the end of the J-domain and the beginning of the glycine- and phenylalanine-rich region. (B) Ribbon diagram of the X-ray crystal structure of the 382-residue amino-terminal fragment of Hsp70 that retains ATPase activity. Glu175 is the conserved residue in Hsp70 that is represented in E coli DnaK (Hsp70) by Glu171. This residue is denoted because it was demonstrated to function in interdomain communication between the ATPase domain and the polypeptide-binding domain of Hsp70 (see text). Glu175 is also located near R167, which is found in an acidic cleft where J-domain binding to Hsp70 is proposed to occur (see text). Gly382 denotes the terminus of ATPase domain. β-Strands are shown in gold, and α-helices are in pink. The J-domain and Hsp70 ATPase fragment structures were from PDB files 1BQZ and 1HJO, respectively, and the images shown were generated with Rasmol