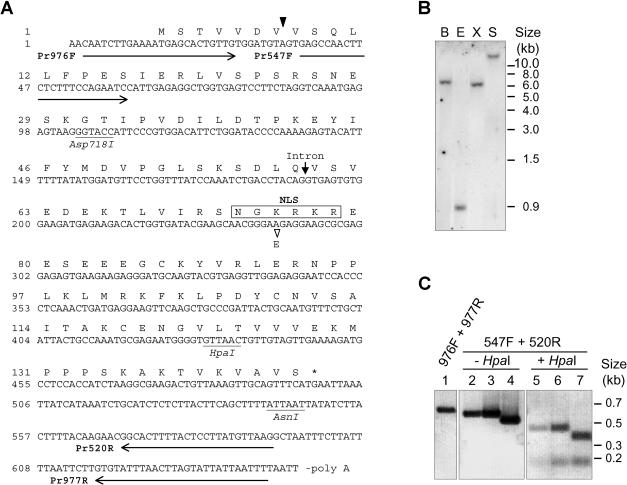
Fig 1.
Structure of tomato Hsp16.1-CIII. (A) The sequence represents the complementary deoxyribonucleic acid (cDNA) clone of LpHsp16.1-CIII including the 5′-random amplification of cDNA ends–derived extension and the open reading frame encoding amino acids 1 to 144. The arrowhead between codon 7 and 8 points to the 5′ end of the cDNA clone rescued by the yeast 2-hybrid screening. The position of the intron insertion at the genomic level is indicated by an arrow. The basic cluster of amino acid residues forming the putative nuclear localization signal, including the amino acid residue exchange K75E, is boxed. Arrows underlining the cDNA sequence correspond to the position and orientation of primers used for the polymerase chain reaction (PCR) analysis shown in (C). (B) Southern blot of genomic DNA from Lycopersicon peruvianum confirms the existence of a single Hsp16.1-CIII gene in tomato. B, BamHI; E, EcoRI; S, SalI; X, XbaI. (C) PCR analysis of genomic DNA (lanes 1 and 2), plasmid DNA containing an Hsp16.1-CIII cDNA clone including the 63-bp intron (MSA2, lanes 3 and 5), or the cDNA clone corresponding to the mature messenger ribonucleic acid of Hsp16.1-CIII (MSA3, lanes 4 and 7). The identity of the PCR products (lanes 2 to 4) was confirmed by restriction analysis using the diagnostic HpaI site (lanes 5–7). Numbers on top refer to primer pairs used for amplification of the corresponding PCR products (for primer sequences see Materials and Methods, and for their positioning in the Hsp16.1-CIII sequence see [A]). Size differences between fragments in lanes 3 and 4 or in lanes 6 and 7 correspond to the short intron of 63 bp