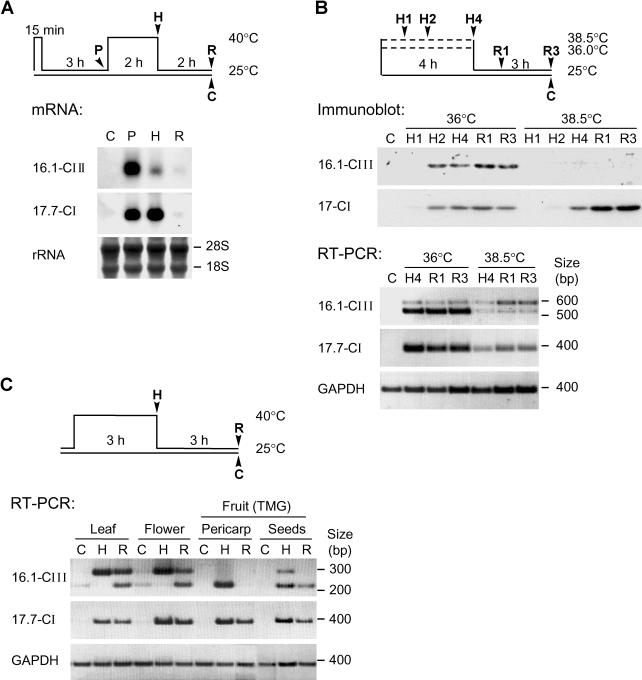
Fig 2.
Heat stress–dependent expression of Hsp16.1-CIII. (A) Tomato suspension culture cells were submitted to the indicated heat stress regime (see pictograph). Ribonucleic acid (RNA) samples were prepared after preinduction (P), heat stress (H), and recovery (R). The RNA was analyzed by Northern blotting using Hsp16.1-CIII– and Hsp17.7-CI–specific probes. Ribosomal RNA staining with methylene blue served as loading control. (B) Analysis of Hsp16.1-CIII expression in tomato suspension culture cells subjected to heat stress treatments at 36°C and 38.5°C (see pictograph on top). For total protein and RNA preparation, aliquots of cells were harvested as indicated, ie, samples H1, H2, and H4 at 1, 2, and 4 hours of heat stress, samples R1 and R3 at 1 and 3 hours of recovery, and sample C represents untreated control cells. The analysis of Hsp17-CI species in the immunoblot detection (above) and Hsp17.7-CI messenger RNA (mRNA) detection by reverse transcriptase–polymerase chain reaction (RT-PCR) (below) was used as positive control for the heat stress effect. The lack of Hsp16.1-CIII accumulation after heat stress treatment at 38.5°C is correlated to an increasing level of its unspliced pre-mRNA in the corresponding RT-PCR samples. For amplification of Hsp16.1-CIII transcripts, primers Pr547F and Pr520R were used (see Fig 1 and Materials and Methods). (C) RT-PCR analysis of Hsp16.1-CIII and Hsp17.7-CI expression in different tomato tissues under control (C), heat stress (H), and recovery (R) conditions. Total RNA was prepared from leaves, flowers, and pericarp and seeds of mature green fruits. In contrast to the RT-PCR analysis shown in part B, primer Pr520R was replaced by Pr548R, which binds complementarily to nucleotides 219–242 of the complementary deoxyribonucleic acid sequence (Fig 1A). As in part B, the glyceraldehyde-3-phosphate dehydrogenase signal was used as internal control for the RT-PCR reaction