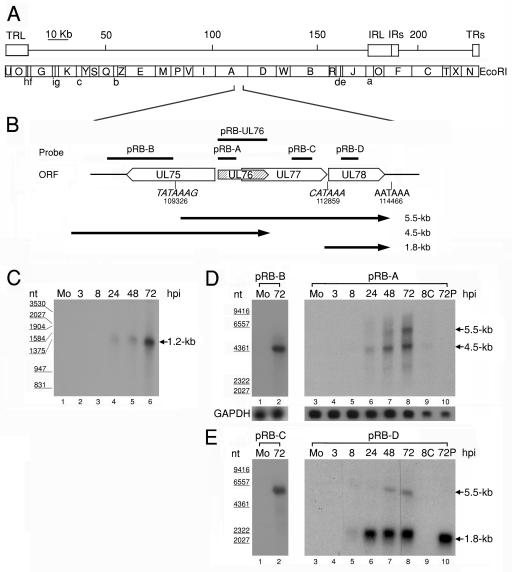
FIG. 1.
(A) Schematic diagram of EcoRI physical maps in the HCMV AD169 genome. The scale indicates the genomic position. The genomic arrangement around UL76 is enlarged in the diagram. (B) Schematic depiction of the genomic region from UL75 to UL78 with the putative transcriptional TATA boxes (nucleotides 109326 and 112859) and the potential poly(A) termination signal (nucleotides 114466). The putative transcripts are indicated by black lines with arrows below the ORFs. Lengths are shown on the right (in kilobases). (C) RNase protection analysis with poly(A)+ RNA purified during the HCMV replication cycle was employed to detect RNA encompassing the UL76 coding sequence. A radiolabeled UL76 antisense transcript complementary to the full-length coding sequence was generated from pRB-UL76 and used for the analysis. Lanes: 1, poly(A)+ RNA from mock-infected cells (Mo); 2 to 6, poly(A)+ RNA harvested at 3, 8, 24, 48, and 72 h postinfection, respectively. The sizes of molecular mass markers are indicated in nucleotides (nt) on the left. Northern blot analyses of UL76 transcriptional expression kinetics during the HCMV replication cycle are shown. Samples of poly(A)+ RNA were electrophoretically separated with a 1% formaldehyde agarose gel. (D) Single-stranded riboprobes specific for UL76 sequence (pRB-A) and the 5′ noncoding sequence of UL76 (pRB-B) were used for hybridization. (E) Northern blots were probed with sequences specific for UL77 (pRB-C) and UL78 (pRB-D). Lanes: 1 and 3, 1 μg of poly(A)+ RNA from mock-infected cells (Mo); 2, 1 μ of poly(A)+ RNA harvested at 72 h postinfection; 3 to 8, 1 μg of poly(A)+ RNA harvested at 3, 8, 24, 48, and 72 h postinfection, respectively; 9, 0.5 μg of poly(A)+ RNA harvested at 8 h postinfection in the presence of cycloheximide (100 μg/ml); 10, 0.5 μg of poly(A)+ RNA harvested at 72 h postinfection in the presence of phosphonoformic acid (150 μg/ml). The transcriptional levels of GAPDH were monitored as an internal control. The sizes of molecular size markers are indicated on the left.