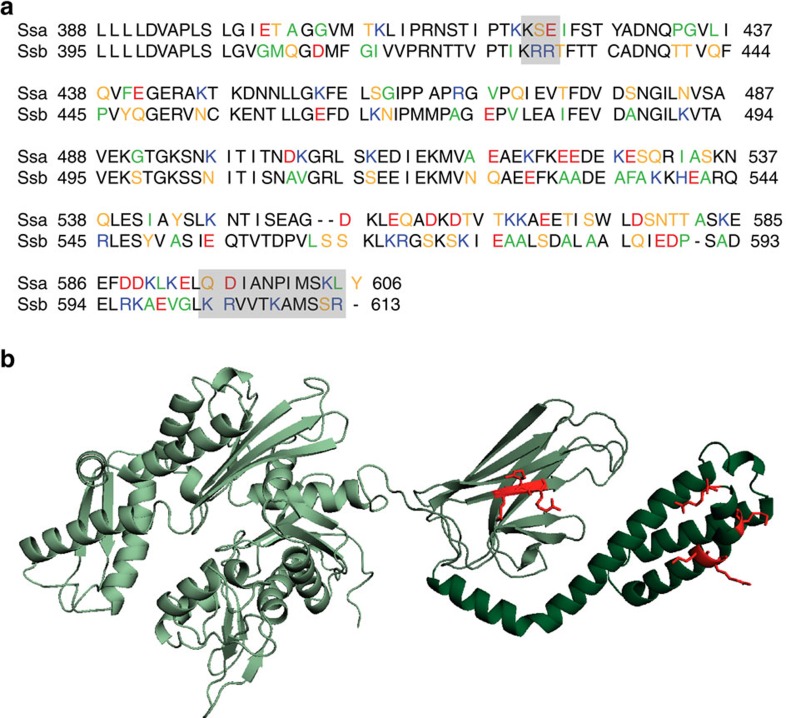
Figure 1. Two positively charged regions that differ between the yeast Hsp70s Ssa and Ssb might be involved in ribosome binding of Ssb.
(a) Alignment of the substrate binding domain (SBD) and the C-terminal domain (CTD) of the yeast Hsp70s Ssa1 (upper lane) and Ssb1 (lower lane). Residues were marked according to their character: neutral/non-polar (green), neutral/polar (yellow), basic (blue), acidic (red); residues in black are identical in Ssa1 and Ssb1 or belong to the same group of amino acids as determined above. Grey boxes mark patches that differ between Ssa1 and Ssb1 and show a basic character in Ssb1. (b) Structural model of Ssb1 based on a DnaK structure (with ADP and substrate;49) showing the N-terminal nucleotide binding domain in light green, the SBD in middle green and the CTD in dark green. Basic residues (K, R) which are marked as grey boxes in a are highlighted in red.