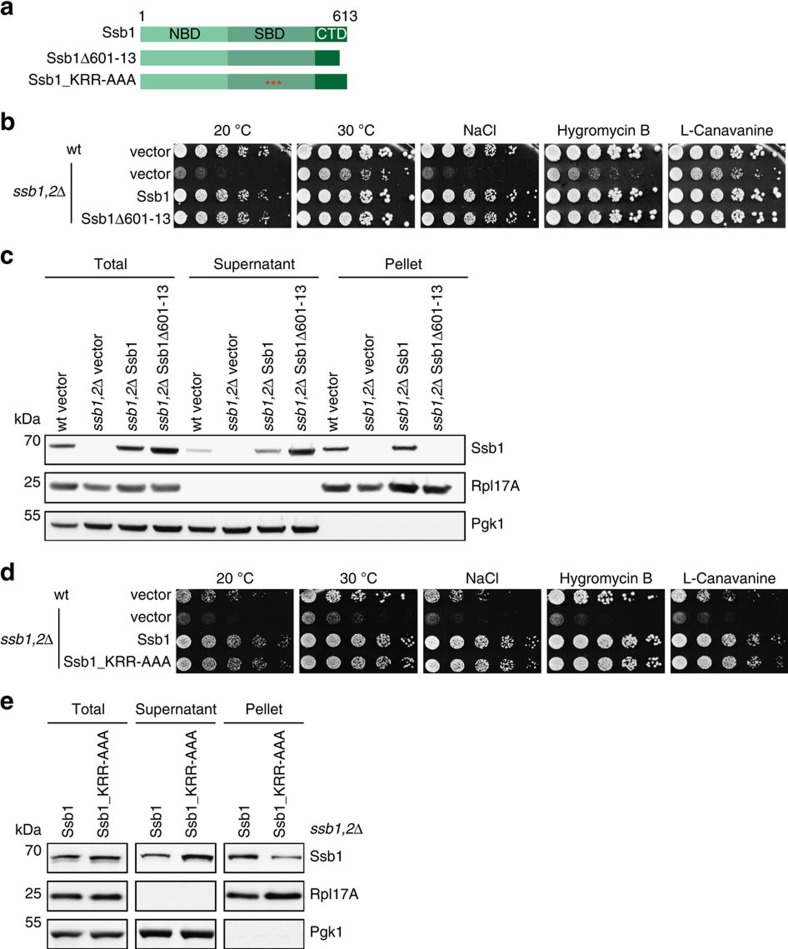
Figure 2. Mutations that influence ribosome binding of Ssb do not hamper growth.
(a) Schematic overview of Ssb domains and the constructs used. Ssb1Δ601–13 lacks 13 C-terminal residues; red asterisks mark point mutations (Ssb1_KRR428AAA). (b) Growth analysis of Ssb C-terminal deletion mutant. Wild type (wt) or ssb1,2Δ cells transformed with either empty vector or different Ssb1 versions were adjusted to OD600=0.4 and spotted in fivefold serial dilutions onto SC-URA plates (that may contain certain additives), which were incubated at 30 °C for two days or at 20 °C for five days. (c) Ribosome sedimentation assay to investigate ribosome binding of Ssb1Δ601–13. Cells were grown to exponential growth phase in SC-URA media and lysates were adjusted to same A260 levels. 1.5 A260 units were loaded onto a 25% (w/v) sucrose cushion followed by ultracentrifugation to pellet ribosomes. Equal amounts of adjusted totals, supernatant and ribosomal pellet fraction were analysed via SDS–PAGE followed by Western blotting. (d) Growth analysis of Ssb1_KRR428AAA mutant as described in b. (e) Ribosome sedimentation assay as described in c to analyse ribosome binding of Ssb1_KRR428AAA mutant.