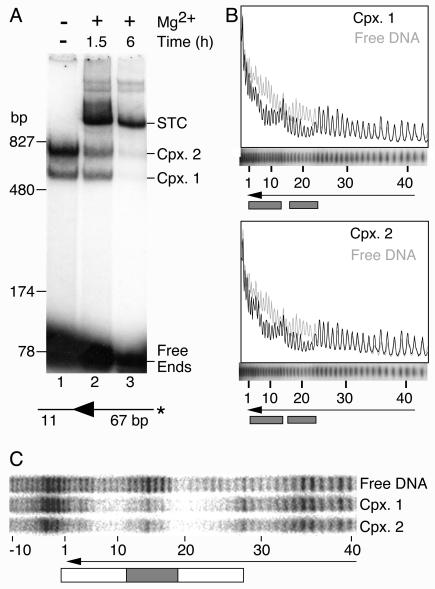
FIG. 4.
Complexes 1 and 2 are probably derived from the Himar1 PEC. (A) Transposition reactions were supplemented with 10 mM MgCl2 to initiate the reaction and were incubated at 30°C for the indicated times. An autoradiogram of an EMSA is shown. The smearing of the STC band at the 1.5-h time point makes it appear to have more counts than in the 6-h time point. However, quantification with a phosphorimager showed that there were 1.2-fold more counts in the STC band at the 6-h time point. The DNA fragment was prepared by digesting pKL97 with PstI-SalI. The complexes were assembled in 15-μl reactions with 60 fmol of DNA and 60 fmol of transposase (4 nM). *, position of radioactive label on the transposon arm. Cpx., complex. (B) Hydroxyl radical footprints. Complexes were assembled with the same DNA fragment as that used for panel A, with the radioactive label on the nontransferred strand. The reactions (20 μl) contained 80 fmol of transposase (4 nM) and 37 fmol of transposon end. The reaction mixture was treated with hydroxyl radicals to generate a footprint as described previously (15, 51). Complex 1, complex 2, and the free DNA from nine such reactions were purified in an EMSA and pooled, and the footprint was displayed on a DNA sequencing gel. Traces were generated with Fuji Image Gauge software. The region of the sequencing gel containing the footprint of the complexes is shown aligned below the traces. The arrowhead shows the position of the transposon end. The numbers of base pairs inside the transposon end are indicated. Shaded boxes indicate regions of hydroxyl radical protection. (C) Copper phenanthroline footprints. The complex assembly reaction (100 μl) contained 100 fmol of transposon end and 600 fmol of transposase (30 nM). The DNA was prepared by digesting pKL97 with XbaI-Acc65I and were labeled on the nontransferred stand by end filling. The reaction was treated with 20 ng of heparin (to reduce the background smear) and loaded into five lanes of an EMSA gel. The complexes were treated with copper phenanthroline in the gel as described previously (42), and the footprints were displayed on a DNA sequencing gel. The open box indicates a region of protection extending from the transposon end to bp +28. The shaded box indicates a strong region of protection centered on bp +14.