Fig. 6.
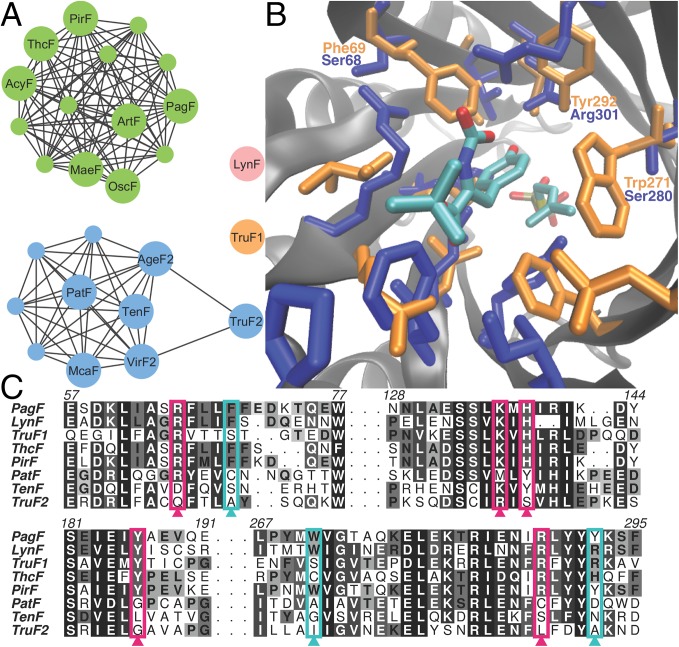
(A) Sequence similarity network showing the clustering of F family enzymes into groupings that reflect their cognate substrate preferences. Using a threshold cutoff E value of 10−80, PagF clusters with other enzymes that prenylate on Tyr (shown as green circles). Members that do not demonstrate activity, such as PatF, cluster and are shown in blue spheres. LynF, which catalyzes reverse prenylation on Tyr (in pink), and TruF1, which catalyzes reverse prenylation on Ser/Thr (in orange), do not cluster with any other members. (B) Structure-based homology model of TruF1 (in blue), superimposed on the structure of PagF (in orange), showing changes in the active site composition that would favor the smaller, polar Ser/Thr over Tyr as a substrate. (C) Multiple sequence alignment of F family enzymes illustrating that inactive members lack at least one residue involved in DMAPP binding (shown in pink), and that TruF1 contains polar amino acids at conserved hydrophobic residues (in cyan).