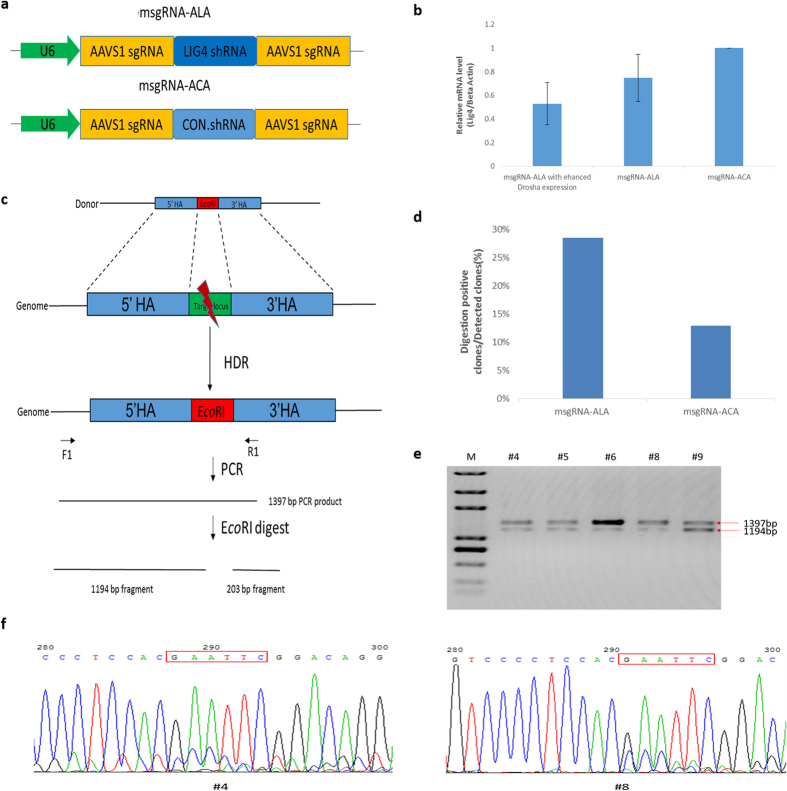
Figure 4. HDR-based precise genome editing assay applying the sgRNA-shRNA structure.
(a) The design of the msgRNA-ALA construct and the control msgRNA-ACA. The LIG4.shRNA and CON.shRNA represent the shRNA designed against LIG4 gene and the control irrelevant shRNA respectively. The two AAVS1.sgRNA scaffolds on the both side of shRNA were designed targeting the same site. (b) The down-regulation of LIG4 gene with msgRNA-ALA. The relative LIG4 mRNA levels (LIG4/Beta-actin) were measured using qRT-PCR. Error bars represent standard deviation, n = 3. (c) An illustration for the HDR-based genome editing of AAVS1 locus and the detection for positive clones. The donor DNA was designed with the PAM motif of the AAVS1 target site replaced by the EcoR I cutting site. This design allows to identify genome edited cell clones by PCR and EcoR I-digesting assay. 1194 bp and 203 bp bands should appear if the genome DNA has been successfully edited by HDR-based repair. (d) Percentage of digestion positive clones within detected clones was calculated to evaluate the precise genome editing efficiency. The PCR and EcoR I-digesting assay demonstrated 10 positive clones out of 35 cell clones (10/35) for the msgRNA-ALA group, while 4 out of 31 clones (4/31) for the msgRNA-ACA control (Supplementary Fig. 3). Three independent replicates were performed for each group. (e) The representative results for the detection of positive clones by EcoR I digestion. The two bands (1397 bp and 1194 bp) indicated that the detected clones were all heterozygotes with one allele edited. This was a cropped gel and the full-length gel images were shown in Supplementary Fig. 4. (f) Representative sequencing results for the PCR products from clones #4 and #8. The lapped peaks indicated that both clones were heterozygotes. The EcoR I site that was introduced is indicated by red rectangles.