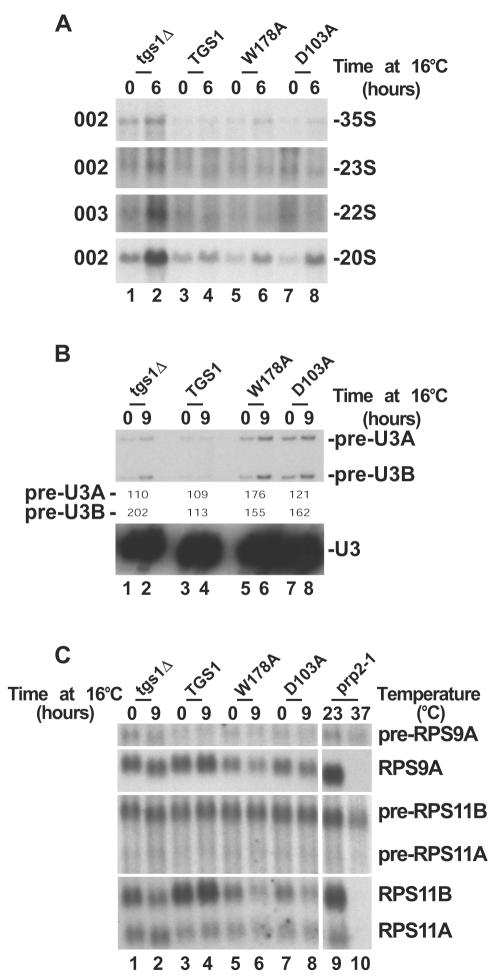
FIG. 3.
Cap trimethylation is not required for pre-rRNA processing. Pre-rRNA processing (A) and splicing analysis (B and C) of Tgs1p catalytic mutants (D103A and W178A), a wild-type isogenic control (TGS1) and a strain with TGS1 deleted (tgs1Δ) grown at 30°C and transferred to 16°C for up to 9 h are shown. (A) Pre-rRNA processing analysis by Northern blotting. The oligonucleotide probes (see Fig. 1A) used are indicated on the left. (B) U3 splicing analysis by primer extension. The splicing defect was estimated by PhosphorImager quantitation (Typhoon; Amersham Biosciences) and expressed as (signal at 16°C/signal at 30°C) × 100. (C) RP pre-mRNA splicing analysis by Northern blotting. A prp2-1 strain grown at 23°C and transferred to 37°C for 90 min was used as a control. See Materials and Methods for the oligonucleotide probes used.