Abstract
Tuberous sclerosis complex (TSC) is a genetic disease caused by a mutation in either the tsc1 or tsc2 tumor suppressor gene. Recent studies have demonstrated that TSC2 displays GAP (GTPase-activating protein) activity specifically towards the small G protein Rheb and inhibits its ability to stimulate the mTOR signaling pathway. Rheb and TSC2 comprise a unique pair of GTPase and GAP, because Rheb has high basal GTP levels and TSC2 does not have the catalytic arginine finger found in Ras-GAP. To investigate the function of TSC2 and Rheb in mTOR signaling, we analyzed the TSC2-stimulated Rheb GTPase activity. We found that Arg15, a residue equivalent to Gly12 in Ras, is important for Rheb to function as a substrate for TSC2 GAP. In addition, we identified asparagine residues essential for TSC2 GAP activity. We demonstrated a novel catalytic mechanism of the TSC2 GAP and Rheb that TSC2 uses a catalytic “asparagine thumb” instead of the arginine finger found in Ras-GAP. Furthermore, we discovered that farnesylation and membrane localization of Rheb is not essential for Rheb to stimulate S6 kinase (S6K) phosphorylation. Analysis of TSC1 binding defective mutants of TSC2 shows that TSC1 is not required for the TSC2 GAP activity but may function as a regulatory component in the TSC1/TSC2 complex. Our data further demonstrate that GAP activity is essential for the cellular function of TSC2 to inhibit S6K phosphorylation.
Tuberous sclerosis complex (TSC) is caused by mutations in either of the tumor suppressor genes tsc1 and tsc2 and is characterized by the development of hamartomas in a variety of tissues (8, 57). The hamartomas found in TSC patients are benign tumors existing in a wide range of tissues including kidney, brain, lung, heart, and skin. The most common clinical complications are seizure and mental retardation, kidney failure, and lung problems due to tumor growth in respective organs. TSC1 and TSC2 form a complex (TSC1/2) in intact cells, and the interaction between TSC1 and TSC2 appears to be important for the stability of both proteins and for their physiological functions (27). It has been well demonstrated that the TSC1/2 complex suppresses cell growth by inhibiting the mammalian target of rapamycin (mTOR) pathway (18, 20, 23, 26, 28, 29, 36, 40, 52). mTOR, a protein kinase, is a central controller of cell growth and stimulates phosphorylation of two key translation regulators, ribosomal S6 kinase (S6K) and eukaryote initiation factor 4E binding protein (4EBP1) (5, 6, 14, 24, 44), whereas TSC1/2 inhibits the phosphorylation of both S6K and 4EBP1 (38). The mechanism of mTOR inhibition by TSC1/2 is not fully understood.
TSC2 contains a region of limited homology to the catalytic domain of the Rap1-GTPase-activating protein (GAP) (16). Both genetic and biochemical studies have demonstrated that Rheb (Ras homolog enriched in brain) is a direct target of TSC2. TSC2 functions as a GAP to inhibit Rheb activity (7, 19, 22, 45, 50, 53, 59). In vitro biochemical studies show that TSC2 can stimulate GTP hydrolysis of purified Rheb. Furthermore, Rheb GTP levels are elevated in TSC2-deficient cells (19), and overexpression of TSC1/2 decreases the GTP levels of cotransfected Rheb. These studies strongly indicate that TSC1/2 inhibits the mTOR/S6K/4EBP1 signaling pathway by stimulating the GTP hydrolysis of Rheb and Rheb functions between TSC2 and mTOR (30, 35).
Rheb is a member of the Ras superfamily GTPases and shares the highest homology with Ras and Rap (55). The Rheb gene is highly conserved in eukaryotes from yeast to mammals (41, 54). Genetic studies of fly and fission yeast indicate that Rheb plays an important role in the stimulation of cell growth and regulation of G0/G1 cell cycle progression (32, 41, 45, 50, 56, 59). The growth arrest phenotype caused by Rheb mutation in Schizosaccharomyces pombe can be complemented by human Rheb (56), suggesting the conservation of Rheb function from yeast to human. The precise physiological functions of Rheb were unknown in high eukaryotes until recently. Both genetic studies in Drosophila melanogaster and biochemical studies in mammalian cells have shown that Rheb is involved in signal transduction pathways that regulate cell growth (7, 19, 22, 45, 50, 53, 59). Homozygous inactivation of Rheb is lethal, while mosaic analyses of Rheb mutant cells in Drosophila show that the inactivation of Rheb decreases cell size. In contrast, overexpression of Rheb increases cell size. Genetic epistatic analysis demonstrates that Rheb functions between TSC1-TSC2 and TOR. In cultured cells, overexpression of Rheb potently stimulates the phosphorylation of S6K and 4EBP1, while downregulation by interference RNA decreased the phosphorylation of S6K and 4EBP1. These data establish Rheb as an important regulator between TSC1/2 and mTOR to stimulate cell growth.
Rheb is unique in that it contains an arginine (Arg15) at the position equivalent to Gly12 in Ras (55). This is significant because Gly12 in Ras is highly conserved in the Ras superfamily, and mutation to any other residue except proline results in constitutive activation of Ras (48). As expected, Rheb contains high basal GTP levels. Interestingly, Ras with a mutation in Gly12 cannot be inactivated by Ras-GAP, while wild-type Rheb can still be inactivated by TSC2 GAP activity. These results suggest that the GTP hydrolysis of Rheb stimulated by the GAP activity of TSC2 is fundamentally different from that of Ras and Ras-GAP.
GAPs from different families do not share obvious sequence homology, although members from any one subfamily do (15). Accordingly, they are termed Ras-GAP, Rap-GAP, and so on. The typical model of GAP catalysis has been well established from studies of the Ras/Ras-GAP (p120 GAP) system. Ras-GAP works by stabilizing the existing catalytic machinery of Ras via supplementing an external arginine residue, termed the arginine finger (47). However, this arginine finger is not conserved in the TSC2 GAP domain. Furthermore, it is still an open question whether the GAP activity is absolutely required for the cellular function of TSC2 to inhibit mTOR signaling. Considering the critical role of Rheb and TSC2 in cell growth regulation and tumor development (3, 17), it is of importance to investigate Rheb GTPase activity as well as the mechanism of how the TSC2 GAP stimulates Rheb GTP hydrolysis. To better understand the mechanism of Rheb GTP hydrolysis and to further establish the functional importance of GAP activity in the physiological function of TSC2, we report here the biochemical and functional characterizations of Rheb GTPase and TSC2 GAP activity. Consistent with the recently reported Rap1-GAP structure (13), we show that TSC2 and Rheb employ a novel mechanism to hydrolyze GTP utilizing an “asparagine thumb.”
MATERIALS AND METHODS
Antibodies and plasmids.
Anti-phospho-S6K(T389) was from Cell Signaling Inc., and anti-Myc antibodies and antihemagglutinin (anti-HA) was purchased from Covance. pcDNA3-HA-TSC1, pcDNA3-HA-TSC2, and pRK5-myc-Rheb constructs were described previously (10, 17). All point mutation constructs of TSC2 and Rheb were generated by using a QuikChange site-directed mutagenesis kit (Stratagene) and verified by DNA sequencing. All deletion constructs of TSC2 were generated by a standard PCR cloning strategy using the pcDNA3-HA-TSC2 construct as a template. The PCR fragments of truncated TSC2 were subcloned in frame into the NotI/XhoI sites of pcDNA3-HA (30).
Cell culture, transfection, and immunoprecipitation.
HEK293 cells were seeded and maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. Transfection was performed in serum-free condition with Lipofectamine reagent (Invitrogen) according to the manufacturer's instructions. Cells were lysed in lysis buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1% Triton X-100, 50 mM NaF, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml) and immunoprecipitated with the indicated antibodies and protein G-Sepharose beads. Immunocomplexes were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting.
Measurement of GTP- and GDP-bound Rheb.
HEK293 cells were cultured in six-well plates and cotransfected with various plasmids with the Lipofectamine reagent. Thirty-six hours after transfection, the cells were washed once with phosphate-free DMEM (DMEM without sodium phosphate and sodium pyruvate, catalog number 11971-025; GIBCO) and incubated with 1 ml of phosphate-free DMEM for 90 min. Cells were then incubated with 25 μCi of [32P]phosphate (ICN)/ml for 4 h. After the labeling, cells were lysed with prechilled lysis buffer (0.5% NP-40, 50 mM Tris [pH 7.5], 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml; 200 μl per well of a six-well plate). To avoid lysing the nuclei, the cells were incubated with lysis buffer for just 30 s with gentle shaking. The lysate was then centrifuged at 12,000 × g for 15 min at 4°C. The supernatant (160 μl) was transferred to a fresh tube. Sixteen microliters of NaCl (500 mM) was added to 160 μl of supernatant to inhibit GAP activity in the lysate. To immunoprecipitate Myc-tagged Rheb, 3 μg of anti-Myc antibody and 10 μl of a protein-G Sepharose bead slurry were added to the supernatant and incubated with gentle rocking for 1 h at 4°C. The beads were washed with wash buffer 1 (50 mM Tris [pH 8.0], 500 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.5% Triton X-100) three times at 4°C. The beads were then washed with wash buffer 2 (50 mM Tris [pH 8.0], 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.1% Triton X-100) three times at 4°C. The Rheb-bound nucleotides were eluted with 20 μl of elution buffer (2 mM EDTA, 0.2% sodium dodecyl sulfate, 1 mM GDP, 1 mM GTP) at 68°C for 10 min. Ten microliters of eluted nucleotides was then applied onto polyethyleneimine cellulose plates (Baker-flex, 20 cm by 20 cm). After applying the sample, the plate was soaked in methanol and dried with a hair dryer. The bottom portion (below the line where the samples are loaded) of the plate was immersed in methanol again, and the plate was placed in a sealed chromatography chamber that was filled with 0.75 M KH2PO4 (pH 3.4) to a depth of 1 cm. The chamber was closed, and the solvent was allowed to ascend to the top of the plate. The plate was then removed and air dried. GTP and GDP resolved by thin-layer chromatography were visualized and quantified by a phosphoimager.
RESULTS
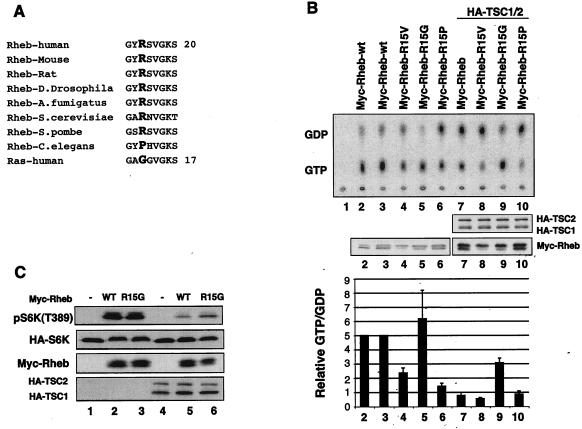
Rheb-R15G is partially resistant to TSC2 GAP.
Rheb has a high basal GTP level that is probably due to an arginine residue at the position corresponding to Gly12 in Ras (21). It has been reported that the replacement of the Arg15 of Rheb by a glycine residue does not decrease Rheb GTP levels as determined by an indirect assay (21). To investigate the functional interaction between Rheb and TSC2, Arg15 in Rheb was mutated to valine (R15V), glycine (R15G), and proline (R15P). These residues were chosen because glycine and valine are found in wild-type and oncogenic Ras, respectively. There are two reasons we mutated Arg15 to a proline residue: first, Pro15 is found in Caenorhabditis elegans Rheb and is the only exception in all identified Rheb homologs (Fig. 1A); second, a mutation of Gly12 in Ras to proline but not any other residue does not activate Ras (48). To determine the nucleotide levels, mutant Rheb was transfected into HEK293 cells, and the transfected cells were labeled with [32P]phosphate. Rheb protein was immunoprecipitated, and the bound nucleotides (GTP and GDP) were separated by thin-layer chromatography and quantified by phosphoimaging. Our data show that wild-type Rheb, Rheb-R15V, Rheb-R15G, and Rheb-R15P contain high GTP levels (from 20 to 66%) (Fig. 1B). It is worth noting that wild-type Ras contains less than 5% GTP under similar experimental conditions (data not shown).
FIG. 1.
Rheb-R15G cannot restore the GTPase activity and is less sensitive to TSC2 GAP activity. (A) Rheb contains an arginine residue (the residue in boldface type) at the position corresponding to codon 12 of Ras, which has a glycine. Numbers indicate positions of the last residues. (B) Mutation of Arg15 affects Rheb GTP binding and sensitivity to TSC2 GAP. Wild-type and mutant Myc-Rheb were transfected into HEK293 cells in the presence or absence of TSC1/2 as indicated and labeled with [32P]phosphate. Myc-Rheb was immunoprecipitated, and the bound nucleotides were eluted and resolved on a cellulose plate. The expression levels of Myc-Rheb and TSC1/2 in a parallel experiment (middle) and quantification of Rheb nucleotide binding (bottom) are shown. The ratio of GTP to GDP was calculated by the following formula: (GTP counts/3)/(GDP counts/2). The values are data from three independent experiments. (C) Rheb-R15G is less sensitive to the TSC1/2 complex. HA-S6K was transfected into HEK293 cells in the presence or absence of Myc-Rheb and TSC1/2. Phosphorylation of S6K was determined by phosphospecific antibody against phospho-S6K(T389). WT, wild type.
The Rheb-R15P mutant displays a basal GTP level significantly lower than that of wild-type Rheb (Fig. 1B). It is interesting that there is no TSC2 homolog in the C. elegans genome. Therefore, one intriguing interpretation is that Pro15 maintains C. elegans Rheb at a low GTP level, thus making GAP activity unnecessary for the C. elegans Rheb. Alternatively, a different C. elegans GAP unrelated to TSC2 may work on its Rheb homolog.
Consistent with previously reported results, Rheb-R15G still has a high GTP level. The mutation of Gly12 in Ras or its equivalent position in Rap results in resistance to inactivation by its GAP (37). To test the effect of TSC2 GAP activity on these Rheb mutants, in vivo labeling experiments were performed with coexpression of TSC1/2 and the Rheb mutants. We observed that TSC1/2 significantly decreased the GTP levels of the wild type, Rheb-R15V, and Rheb-R15P (Fig. 1B). These data indicate that TSC2 can still stimulate the GTP hydrolysis of Rheb-R15V, while a similar mutation in the oncogenic Ras-G12V is resistant to inactivation by Ras-GAP. Surprisingly, Rheb-R15G was partially resistant to inactivation by TSC1/2 (Fig. 1B). To further test the functional relationship between Rheb-R15G and TSC1/2, we next determined whether Rheb-R15G-induced phosphorylation of S6K is less sensitive to inhibition by TSC1/2 coexpression. Our data showed that Rheb-R15G is reproducibly more resistant to inhibition by TSC1/2 than the wild type as determined by the S6K phosphorylation assay, an indirect functional assay for Rheb (Fig. 1C).
Rheb-Q64L is sensitive to TSC2 GAP.
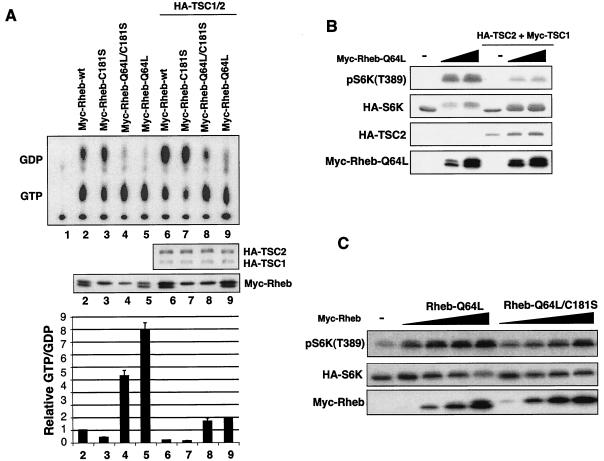
Gln61 of Ras is a catalytic residue important for GTP hydrolysis and is the most frequently mutated residue in human cancer (46). Ras-Q61L has a dramatically increased basal GTP level, and the GTPase activity of Ras-Q61L is resistant to Ras-GAP stimulation (46). We observed that Rheb-Q64L also displays a higher basal GTP level (Fig. 2A). These data are consistent with our previous observation that Rheb-Q64L is more active than wild-type Rheb in stimulating S6K (22). Interestingly, the replacement of Gln64 by leucine in the Rheb GTPase does not abolish the effect of TSC2 GAP. Cotransfection of TSC1/2 significantly increased the GDP level of Rheb-Q64L (Fig. 2A). To further support the above observation, phosphorylation of S6K was determined. We observed that TSC1/2 inhibited the ability of Rheb-Q64L to stimulate S6K phosphorylation (Fig. 2B). These results are completely consistent with the GTP binding data in Fig. 2A. Therefore, TSC2 can still enhance GTP hydrolysis of Rheb-Q64L. Our data suggest that the relationship between Rheb and TSC2 GAP is different from that of Ras and Ras-GAP. TSC2 shares limited sequence homology with Rap-GAP. The three-dimensional structure of Rap1-GAP has recently been reported (13). Our results are completely consistent with the catalytic model proposed for Rap1 and Rap1-GAP in which the glutamine equivalent to Glu61 in Ras is not required for GTP hydrolysis as we observed with Rheb.
FIG. 2.
Farnesylation of Rheb is important for GTP loading but not essential to activate downstream signaling. (A) Nucleotide binding of Rheb mutants. Expression levels of Rheb mutants and TSC1/2 are determined by Western blotting (middle). Quantification of Rheb nucleotide binding is also shown (bottom). wt, wild type. (B) TSC1/2 inhibits Rheb-Q64L mutant-induced phosphorylation of S6K. (C) Rheb-Q64L/C181S is less active to stimulate S6K but does not function as a dominant negative.
Farnesylation is not required for Rheb to activate downstream targets.
Farnesylation of Ras is essential for its membrane localization and biological function (11, 58). The yeast and mammalian Rheb proteins have been shown to be farnesylated (10, 56). To investigate the effect of Rheb farnesylation on its GTPase activity, the cysteine (Cys181) within the farnesylation CaaX motif was substituted by a serine (C181S). Our in vivo labeling experiments show that Rheb-C181S displays a significantly lower GTP-bound level than the wild-type (Fig. 2A), suggesting that membrane localization of Rheb plays a role for Rheb to efficiently load GTP. This observation indicates that the putative guanine nucleotide exchange factor (GEF) may exist in the membrane vicinity if there is a Rheb GEF. However, coexpression of TSC1/2 can still decrease the GTP levels of Rheb-C181S. We found that Rheb-C181S can still activate S6K, although less effectively than wild-type Rheb (data not shown). Therefore, membrane targeting is not essential for Rheb to serve as a substrate for TSC2 nor is it required for Rheb to stimulate S6K. In contrast, Ras-C186S is completely inactive to stimulate the mitogen-activated protein kinase pathway.
Ras-Q61L/C186S is a cytosol-localized GTP-bound dominant-negative mutant. Ras-Q61L/C186S likely prevents membrane recruitment of Ras targets, such as Raf, by sequestering Raf in the cytoplasm. The membrane localization of Raf is essential for Raf activation. To further investigate the importance of Rheb membrane localization in Rheb signaling, we performed experiments with Rheb-Q64L/C181S, a mutant equivalent to Ras-Q61L/C186S, to test the importance of Rheb membrane association. Our data indicate that Rheb-Q64L/C181S does not inhibit S6K phosphorylation. On the contrary, Rheb-Q64L/C181S activates S6K, albeit less potently than does Rheb-Q64L (Fig. 2C). These observations demonstrate that membrane localization is not essential for Rheb function and further support that unlike Ras targets, activation of Rheb downstream targets does not have to occur at the membrane.
Rheb-S20N, Rheb-D60V, and Rheb-D60K do not act as dominant negatives.
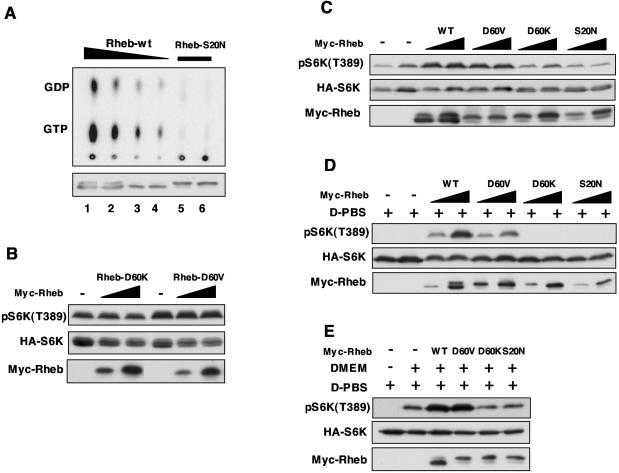
Dominant-negative GTPase mutants are important tools used for elucidating the biological function of a particular GTPase. It has been well demonstrated that the mutation of Ser17 to asparagine in Ras family GTPases creates a dominant-negative mutant which can inhibit endogenous Ras activation by sequestering GEFs (9). We attempted to create a dominant-negative Rheb. Rheb-S20N, which is analogous to Ras-S17N, binds less guanine nucleotides than does wild-type Rheb (Fig. 3A), a property similar to that of Ras-S17N. The nucleotides bound in Rheb-S20N represent less than 1% of the wild-type Rheb. We tested the effect of Rheb-S20N on the phosphorylation of S6K, which depends on Rheb activity. However, coexpression of Rheb-S20N had no significant effect on S6K phosphorylation (Fig. 3B to E). These observations can be explained if Rheb-S20N does not function as a dominant negative. An alternative explanation is that Rheb is not required for the basal S6K phosphorylation. We favor the former possibility (see Discussion).
FIG. 3.
Characterizations of potential dominant-negative Rheb mutants. (A) Rheb-S20N is defective in nucleotide binding. Wild-type Rheb (Rheb-wt) and Rheb-S20N were transfected into HEK293 cells. The transfected cells were labeled with [32P]phosphate. The bound nucleotides (top) of immunoprecipitated Rheb (bottom) are shown. (B) Rheb-D60V and Rheb-D60K do not inhibit S6K phosphorylation. HA-S6K was cotransfected with Rheb mutants in HEK293 cells. Thirty minutes before harvesting, cells were stimulated with fresh DMEM with 10% fetal bovine serum. Phosphorylation of S6K was determined by Western blotting. (C) Rheb-D60V, RhebD60K, and Rheb-S20N do not function as dominant negatives. Experiments are similar to those described above (B), except that cells were not stimulated by fresh medium. (D) Rheb-D60V activates S6K. The transfected HEK293 cells were treated with D-PBS (PBS containing 45 mM glucose) for 30 min before harvesting. (E) Rheb-D60V and Rheb-D60K do not block S6K activation by DMEM. The transfected HEK293 cells were treated with D-PBS for 30 min followed by treatment with DMEM (as indicated) without serum for 30 min before harvesting. WT, wild type.
It was reported that Rheb-D60K and Rheb-D60V act as dominant negatives (51). We performed similar experiments with Rheb-D60K and Rheb-D60V and found that neither Rheb-D60K nor Rheb-D60V blocked S6K phosphorylation (Fig. 3B to E). In contrast, under normal growth conditions (1[1/2]-day culture with no change of the culture medium), Rheb-D60V activated S6K, while Rheb-D60K neither activated nor inhibited S6K (Fig. 3C). We also examined S6K phosphorylation in response to stimulation by fresh medium containing 10% fetal bovine serum. Coexpression of Rheb-D60K or Rheb-D60V did not show any significant effect on S6K phosphorylation in fresh medium (Fig. 3B). To further confirm the stimulatory effect of Rheb-D60V, transfected HEK293 cells were subjected to treatment with Dulbecco's phosphate-buffered saline (D-PBS), which has been well documented to induce S6K dephosphorylation. Rheb-D60V activated S6K, although less efficiently than did wild-type Rheb, while Rheb-D60K and Rheb-S20N did not activate S6K in D-PBS (Fig. 3D).
Replacement of D-PBS by DMEM without serum could activate S6K (Fig. 3E), while the addition of serum alone in PBS did not activate S6K (data not shown). We also examined the effects of Rheb mutants on DMEM-stimulated S6K phosphorylation. Our data show that neither Rheb-D60K nor Rheb-S20N blocked the DMEM-induced S6K phosphorylation (Fig. 3E). Under the same conditions, wild-type Rheb or Rheb-D60V activated S6K. The experiments described above clearly demonstrate that Rheb-D60V and Rheb-D60K do not function as dominant negatives while Rheb-D60V is active to stimulate S6K phosphorylation under several conditions tested. Our data are inconsistent with results described by Tabancay et al., who reported the dominant effect of Rheb-D60K and Rheb-D60V. One possible explanation could be the difference in levels of expression between the two studies. However, it is worth noting that the expression levels of the Rheb mutants in our experiments were high.
TSC1 is not required for TSC2 GAP activity towards Rheb.
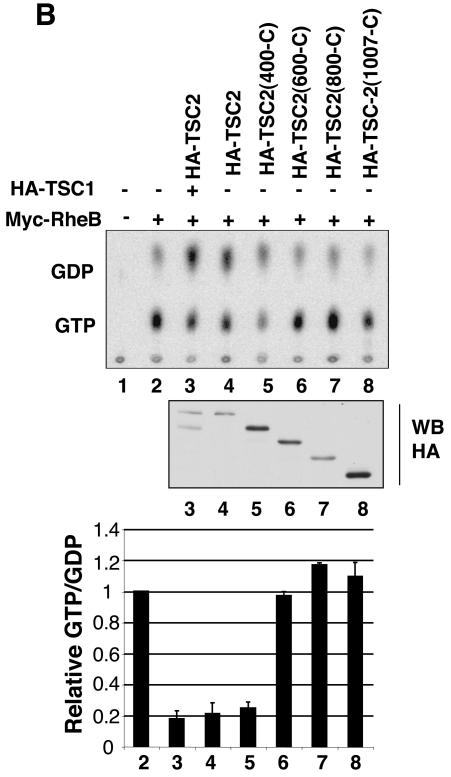
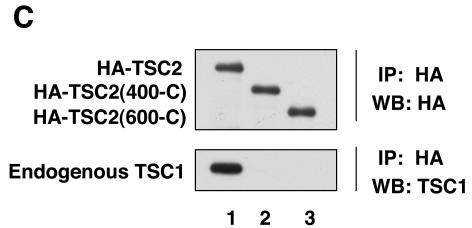
TSC1 and TSC2 form a stable physical complex, which is important for the functions of these two tumor suppressor gene products (23, 42). The requirement of TSC1 for TSC2 GAP activity is not clear from a review of the literature (7, 19, 22, 53, 59). When same quantity of TSC2 plasmid was used in transfection, coexpression of TSC1 caused a significant increase in the reduction of Rheb GTP levels than did the transfection of TSC2 alone. However, the effect of TSC1 is complex because TSC1 coexpression significantly increased the TSC2 protein level. To avoid this complication, different amounts of TSC2 plasmid were used to achieve a similar level of TSC2 expression in either the presence or absence of TSC1. We observed that expression of TSC2 alone inhibits Rheb GTP levels, similar to that induced by the TSC1 and TSC2 coexpression if the TSC2 expression level is similar (Fig. 4B). These results would suggest that TSC1 is not required for the GAP activity towards Rheb. However, this interpretation is complicated by the fact that transfection of TSC2 in HEK293 cells may elevate endogenous TSC1 protein levels (39). To overcome this problem, we used a TSC2 mutant with a deletion of the N-terminal region, which is required for interaction with TSC1. It has been reported that amino acids (1 to 418) of TSC2 are responsible for the interaction with TSC1 by yeast two-hybrid assay (Fig. 4A) (27). We performed coimmunoprecipitation to confirm whether TSC2(400-C) can interact with TSC1. Here, we present biochemical data that a TSC2(400-C) mutant does not bind endogenous TSC1 in mammalian cells while wild-type TSC2 coimmunoprecipitated with endogenous TSC1 (Fig. 4C), confirming that the N-terminal region of TSC2 is required for interaction with TSC1. The effect of TSC2(400-C) on Rheb GTP levels was examined. We observed that the expression of TSC2(400-C) efficiently decreased the Rheb GTP level (Fig. 4B). Our data strongly indicate that TSC1 is not required for the GAP activity of TSC2.
FIG. 4.
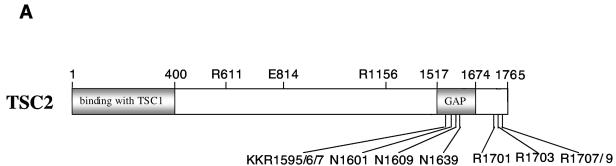
TSC1 is not required for TSC2 GAP activity. (A) Schematic diagram of TSC2. The amino acids mutated in this study are indicated. GAP denotes the region of homology to the catalytic domain of Rap1-GAP. (B) The TSC1 interaction-defective mutant TSC2(400-C) displays GAP activity. TSC2(400-C), TSC2(600-C), TSC2(800-C), and TSC2(1007-C), TSC2 mutants with deletions of the N-terminal 400, 600, 800, and 1,007 residues, respectively. The bound nucleotides (top), expression levels of the TSC2 (middle), and quantification of the Rheb GTP-to-GDP ratio (bottom) are shown. WB, Western blot. (C) The N-terminal region of TSC2 is required for interaction with TSC1. HA-TSC2 and deletion mutants were transfected into HEK293 cells. Cell lysates were immunoprecipitated (IP) by anti-HA antibody for HA-TSC2 and blotted with anti-HA and anti-TSC1.
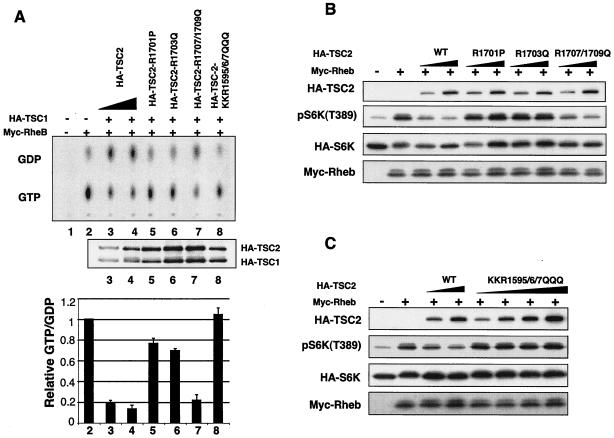
The KKR(1595/6/7), Arg1701, and Arg1703 of TSC2 are essential for TSC2 GAP activity.
The sequence homologous to Rap-GAP occupies only a small part of the TSC2 protein in its C-terminal region (residues 1517 to 1674) (Fig. 4A). Our deletion experiments show that sequence outside of the GAP domain is also important for GAP activity (Fig. 4B). For example, the deletion of 600 amino acid residues at the N terminus completely abolishes the GAP activity of TSC2. The loss of GAP activity is unlikely due to a global alteration of the protein structure because the TSC2 deletions can still be phosphorylated by pathways that regulate TSC2 and these mutants retain their ability to bind the 14-3-3 protein (31).
The biochemical and structural study of Ras-GAP and Rho-GAP reveal two highly conserved arginines essential for GAP activity (47). However, the first arginine, which functions as the catalytic arginine finger in Ras-GAP, is not conserved in TSC2 homologs. Corresponding to the second conserved arginine in Ras-GAP, there are three positively charged residues of KKR at positions 1595, 1596, and 1597 [KKR(1595/6/7)]. Moreover, there is no additional invariant arginine in the TSC2 domain homologous to Rap-GAP. In contrast, four invariant arginines exist in TSC2 that are C terminal to the GAP domain (Arg1701, Arg1703, Arg1707, and Arg1709) (Fig. 4A). In order to determine potential catalytic residues necessary for TSC2 GAP activity, we mutated all of the above-mentioned residues. These mutants are denoted R1701P, R1703Q, double mutant R1707/1709Q, and triple mutant KKR(1595/6/7)QQQ. Mutation at KKR(1595/6/7), Arg1701, and Arg1703 abolished the GAP activity of TSC2, while mutation at Arg1707 and Arg1709 had no significant effect (Fig. 5A). We also examined the ability of these TSC2 mutants to inhibit S6K phosphorylation. Consistently, mutant R1707/1709Q retains the ability to inhibit S6K phosphorylation, and the mutants KKR(1595/6/7)QQQ, Arg1701, and Arg1703 with abolished GAP activity also lost the ability to inhibit S6K (Fig. 5B and C). Our data indicate the importance of these residues in TSC2 function and GAP activity. Interestingly, there are deletion and point mutations at Arg1701 and Arg1703 identified in TSC patients (12, 25), suggesting that the loss of these two arginines abolishes the GAP activity, thereby causing TSC. The above-mentioned data support a functional importance of GAP activity for TSC2 function.
FIG. 5.
Identification of arginines essential for TSC2 GAP activity. (A) The GTP-to-GDP ratio of Rheb was measured in the presence of wild-type and mutant TSC2 as indicated. The expression levels of TSC1 and TSC2 (middle) and quantification of GAP activity of TSC2 point mutations (bottom) are shown. (B) Inhibition of S6K by TSC2 arginine mutants. HA-S6K and wild-type (WT) or mutant TSC2 were cotransfected into HEK293 as indicated. Phosphorylation of S6K was determined by phosphospecific antibody against phospho-pS6K(T389). (C) Inhibition of S6K by the TSC2 wild type and mutants. Experiments were similar to those described above (B).
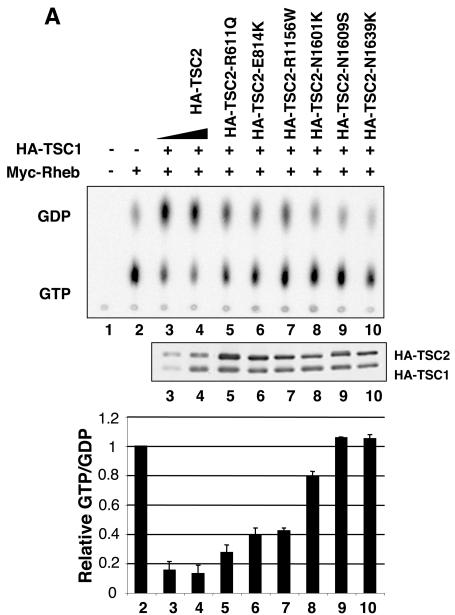
TSC2 disease-related mutants have lower GAP activity.
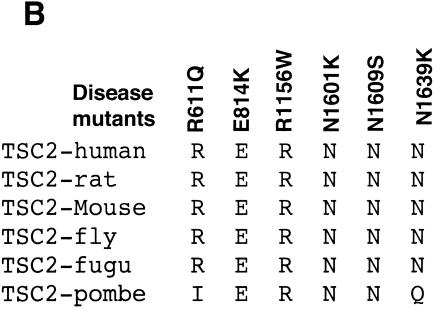
To further reveal the relationship between GAP activity and the physiological function of TSC2, additional disease-derived TSC2 mutants were investigated. We focused on point mutations involving hydrophilic or charged residues, which are presumably localized on the surface of the TSC2 protein (13, 25). We found that three point mutations (N1601K, N1609S, and N1639K) in the GAP domain eliminated GAP activity (Fig. 4A and 6A). Interestingly, all three asparagines are highly conserved in TSC2 (Fig. 6B). The mutant of N1601K has been reported to abolish TSC2 GAP activity (19, 59). Our studies identified three asparagine residues essential for GAP activity. The recently reported three-dimensional structure of Rap1-GAP indicates a catalytic active-site asparagine thumb (13). Interestingly, N1601 corresponds to the position equivalent to the asparagine thumb in Rap1-GAP. Our data suggest that N1601 is the active-site asparagine thumb in the GAP activity of TSC2. Furthermore, the GAP activity of mutants (R611Q, E814, and R1156W) was significantly compromised but not eliminated (Fig. 6A). These results support a notion that GAP activity of TSC2 is important for its physiological function.
FIG. 6.
Identification of GAP activity-essential asparagines corresponding to mutations found in TSC patients. (A) Asparagine residues N1601, N1609, and N1639 are essential for TSC2 GAP activity. The GTP-to-GDP ratio of Rheb was measured in the presence of wild-type and mutant TSC2 as indicated. (B) Alignment of TSC2 mutated residues described above (A). fugu, Fugu rubripes; pombe, S. pombe.
DISCUSSION
Recent studies from several groups, including ours, have established that TSC2, which is responsible for approximately 60% of TSC disease, has GAP activity specifically towards Rheb. There is a Rap1-GAP homology domain in the C-terminal region of TSC2 (Fig. 3A) (16, 34). The GAP domain is the sole functionally recognizable domain in TSC2. Importantly, frequent point mutations in the TSC2 GAP domain are found in TSC patients, indicating the importance of the GAP domain in TSC2 function (1, 25, 33).
Sequence analyses of Rheb and TSC2 suggest that they comprise a pair of atypical small GTPase and GAP. Rheb has an arginine at the position equivalent to Gly12 of Ras, and the arginine finger in Ras-GAP is not conserved in TSC2. Our studies of the biochemical properties of Rheb mutants further imply that the mechanisms of Rheb GTP hydrolysis and stimulation by TSC2 GAP are different from those of Ras based on the following observations. (i) Rheb-R15 (wild type) is a better substrate than Rheb-R15G for TSC2 GAP activity. Rheb-Q64L is still sensitive to TSC2 GAP activity. These observations are in clear contrast to Ras. Ras-G12 (wild type) is sensitive to Ras-GAP, while Ras-G12R is resistant to Ras-GAP. Furthermore, the Ras-Q61L mutation is resistant to GTP hydrolysis stimulation by Ras-GAP. (ii) The mutation of Arg15 to Gly15 in Rheb does not decrease basal GTP level. Moreover, the mutation of Arg15 to Val15 and Pro15 significantly decreases its GTP-to-GDP ratio. In the Ras protein, Ras-G12V and Ras-G12R have higher basal GTP levels than Ras-G12 (wild type). (iii) Rheb-S20N does not function as a dominant-negative mutant although Rheb-S20N does not bind the guanine nucleotide. Ras-S17N functions as a dominant negative by sequestering the upstream activator GEF. Therefore, Rheb displays many unique properties distinct from those of other Ras family members.
The GAP homologous domain of TSC2 (approximately 150 amino acid residues) shares 30% sequence identity with the GAP domain of Rap1-GAP. During the preparation of this paper, the crystal structure of Rap1-GAP was reported (13). Our results are completely consistent with catalytic mechanisms proposed by Daumke et al. for Rap1-GAP. Similar to TSC2, Rap1-GAP does not use an arginine finger as a catalytic residue to stimulate GTP hydrolysis. Instead, Rap1-GAP utilizes an asparagine, termed the asparagine thumb, as the active site to stimulate GTP hydrolysis. This active-site asparagine in Rap1-GAP corresponds to N1601 in TSC2, which is mutated in human disease and is also required for TSC2 GAP activity (Fig. 5). Therefore, we propose that N1601 is the active-site residue asparagine thumb for TSC2 to stimulate Rheb GTP hydrolysis. Our data showing that TSC2 can stimulate GTP hydrolysis of Rheb-Q64L are also consistent with the data observed for Rap1-GAP and Rap1, in which the Ras Q61-equivalent residue is not required for the stimulation of Rap1 GTP hydrolysis by the Rap1-GAP (4). Therefore, our study demonstrates that the catalytic mechanism of TSC2 and Rheb is similar to Rap1-GAP and Rap but is completely different from Ras-GAP and Ras.
Deletion analysis reveals that TSC2 requires sequences outside the GAP domain (residues 1517 to 1674) for GAP activity. Deletion of the N-terminal 600 residues completely abolishes TSC2 GAP activity, suggesting that the GAP domain of TSC2 is larger than that predicted by sequence homology. Our mutational analysis identified two arginine residues, Arg1701 and Arg1703, important for GAP activity. These two residues are located outside of the predicted GAP domain and are also mutated in TSC patients. These two arginine residues are conserved in Rap1-GAP and have been implicated in substrate binding. In addition, mutation of the conserved KKR(1567/8/9) completely eliminated GAP activity. Daumke et al. reported that the mutation of K285 in Rap1-GAP, which corresponds to K1568 in TSC2, abolished GAP activity (4, 13). Based on the three-dimensional structure and binding data, K285 is important to position the catalytic α-helix 7 and is important for substrate binding. Our data are consistent with a similar role for K1568 in TSC2.
We observed a tight correlation between GAP activity of TSC2 and its ability to inhibit S6K phosphorylation. Any mutation with a low TSC2 GAP activity concomitantly decreases its function to inhibit mTOR signaling. These data further demonstrate that the GAP activity of TSC2 is important for its physiological function to regulate protein synthesis and cell growth.
There are two types of dominant-negative Ras mutants. Ras-S17N functions as a dominant negative by sequestering upstream GEF. In contrast, Ras-Q61L/C186S, which is not associated with the membrane but can bind Raf, functions as a dominant negative through binding Raf and preventing Raf activation because Raf activation occurs at the membrane (49). We found that neither Rheb-S20N nor Rheb-Q64L/C181S functions as a dominant negative to inhibit S6K phosphorylation. The soluble Rheb-Q61L/C181S, which presumably can still bind to as-yet-unidentified effectors, can also stimulate S6K phosphorylation. Similarly, Rheb-C181S can also stimulate S6K activation, albeit less effectively. Phosphorylation of S6K was used in our study as the functional assay for Rheb. These results indicate that the activation of Rheb targets may not be restricted to the membrane although direct downstream targets of Rheb have not been identified. A less interesting explanation is that S6K phosphorylation does not require Rheb function, while Rheb-S20N and Rheb-Q61L/C181S do function as dominant negatives by inhibiting activation of endogenous Rheb and its downstream effectors, respectively. However, this explanation is unlikely, based on current genetic and biochemical data that S6K phosphorylation is a real physiological readout of Rheb function (30). First, mutation of Rheb decreases S6K phosphorylation. Downregulation of Rheb by interference RNA also inhibits S6K phosphorylation. Second, the ability of nutrients and insulin to stimulate S6K is abolished in cells containing mutant Rheb. Third, overexpression of Rheb stimulates S6K. Fourth, both GTP binding and the effector domain of Rheb are required for Rheb to stimulate S6K phosphorylation. Furthermore, expression of TSC2, a Rheb GAP, decreases S6K phosphorylation. Together, these observations demonstrate an obligatory function of Rheb in S6K phosphorylation which is a bona fide physiological readout of Rheb, similar to ERK phosphorylation as a physiological readout of Ras.
Consistent with the farnesylation-defective mutants of Rheb having compromised ability to stimulate S6K activity (7, 53), the Rheb-C181S mutant also shows a lower GTP level (Fig. 2A). Because a mutation in Cys181 does not change the sequence involved in GTP binding and hydrolysis, the decrease of the GTP-to-GDP ratio is likely due to sensitivity toward either a GEF or a GAP. However, the latter possibility was ruled out because the sensitivity of Rheb-C181S to TSC2 inhibition was similar to that of the wild type (Fig. 2A). It has been shown that prenylation of Ras proteins is required for efficient stimulation by its GEF (43). It is possible that farnesylation of Rheb is important for Rheb stimulation by GEF, although a Rheb GEF has not been identified. Future studies to investigate the existence and identity of Rheb GEF will be of high significance.
The inactivation of TSC1 or TSC2 causes similar phenotypes, suggesting that they may affect the same downstream targets. For instance, genetic studies show that a mutation in either TSC1 or TSC2 elevates S6K activity (27). However, it remains unclear how or whether TSC1 contributes to TSC2's GAP activity. Studies from Zhang et al., Garami et al., and Tee et al. showed that TSC1 is required for TSC2 GAP activity towards Rheb (19, 53, 59), while studies from Castro et al. (7) and our group (22) showed that TSC1 is not required. Here, we further address this issue by testing the GAP activity of various truncated TSC2 mutants. Our data clearly show that TSC2(400-C), which cannot bind TSC1, still has GAP activity (Fig. 4B), suggesting that TSC2 alone is sufficient to promote GTP hydrolysis of Rheb. However, it has been well demonstrated that TSC1 can bind TSC2 and stabilize TSC2 by preventing degradation (2). Furthermore, TSC1 has been implicated in modulating the subcellular localization of TSC2 (39). Therefore, we propose that TSC1 is not required for Rheb-GAP activity per se but likely plays an important role in regulating the physiological function of TSC2 by modulating the protein stability and localization.
Acknowledgments
We thank H. Chong, C.-H. Lee, and M. Corradetti for critical reading of the manuscript.
This work was supported by grants from the National Institutes of Health and the Walther Cancer Institute.
REFERENCES
- 1.Au, K. S., J. A. Rodriguez, J. L. Finch, K. A. Volcik, E. S. Roach, M. R. Delgado, E. Rodriguez, Jr., and H. Northrup. 1998. Germ-line mutational analysis of the TSC2 gene in 90 tuberous-sclerosis patients. Am. J. Hum. Genet. 62:286-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benvenuto, G., S. Li, S. J. Brown, R. Braverman, W. C. Vass, J. P. Cheadle, D. J. Halley, J. R. Sampson, R. Wienecke, and J. E. DeClue. 2000. The tuberous sclerosis-1 (TSC1) gene product hamartin suppresses cell growth and augments the expression of the TSC2 product tuberin by inhibiting its ubiquitination. Oncogene 19:6306-6316. [DOI] [PubMed] [Google Scholar]
- 3.Bjornsti, M. A., and P. J. Houghton. 2004. The TOR pathway: a target for cancer therapy. Nat. Rev. Cancer 4:335-348. [DOI] [PubMed] [Google Scholar]
- 4.Brinkmann, T., O. Daumke, U. Herbrand, D. Kuhlmann, P. Stege, M. R. Ahmadian, and A. Wittinghofer. 2002. Rap-specific GTPase activating protein follows an alternative mechanism. J. Biol. Chem. 277:12525-12531. [DOI] [PubMed] [Google Scholar]
- 5.Brown, E. J., P. A. Beal, C. T. Keith, J. Chen, T. B. Shin, and S. L. Schreiber. 1995. Control of p70 S6 kinase by kinase activity of FRAP in vivo. Nature 377:441-446. [DOI] [PubMed] [Google Scholar]
- 6.Brunn, G. J., C. C. Hudson, A. Sekulic, J. M. Williams, H. Hosoi, P. J. Houghton, J. C. Lawrence, Jr., and R. T. Abraham. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277:99-101. [DOI] [PubMed] [Google Scholar]
- 7.Castro, A. F., J. F. Rebhun, G. G. Clark, and L. A. Quilliam. 2003. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J. Biol. Chem. 278:32493-32496. [DOI] [PubMed] [Google Scholar]
- 8.Cheadle, J. P., M. P. Reeve, J. R. Sampson, and D. J. Kwiatkowski. 2000. Molecular genetic advances in tuberous sclerosis. Hum. Genet. 107:97-114. [DOI] [PubMed] [Google Scholar]
- 9.Cherfils, J., and P. Chardin. 1999. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci. 24:306-311. [DOI] [PubMed] [Google Scholar]
- 10.Clark, G. J., M. S. Kinch, K. Rogers-Graham, S. M. Sebti, A. D. Hamilton, and C. J. Der. 1997. The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. J. Biol. Chem. 272:10608-10615. [DOI] [PubMed] [Google Scholar]
- 11.Cox, A. D., and C. J. Der. 2002. Farnesyltransferase inhibitors: promises and realities. Curr. Opin. Pharmacol. 2:388-393. [DOI] [PubMed] [Google Scholar]
- 12.Dabora, S. L., S. Jozwiak, D. N. Franz, P. S. Roberts, A. Nieto, J. Chung, Y. S. Choy, M. P. Reeve, E. Thiele, J. C. Egelhoff, J. Kasprzyk-Obara, D. Domanska-Pakiela, and D. J. Kwiatkowski. 2001. Mutational analysis in a cohort of 224 tuberous sclerosis patients indicates increased severity of TSC2, compared with TSC1, disease in multiple organs. Am. J. Hum. Genet. 68:64-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daumke, O., M. Weyand, P. P. Chakrabarti, I. R. Vetter, and A. Wittinghofer. 2004. The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature 429:197-201. [DOI] [PubMed] [Google Scholar]
- 14.Dennis, P. B., S. Fumagalli, and G. Thomas. 1999. Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr. Opin. Genet. Dev. 9:49-54. [DOI] [PubMed] [Google Scholar]
- 15.Donovan, S., K. M. Shannon, and G. Bollag. 2002. GTPase activating proteins: critical regulators of intracellular signaling. Biochim. Biophys. Acta 1602:23-45. [DOI] [PubMed] [Google Scholar]
- 16.European Chromosome 16 Tuberous Sclerosis Consortium. 1993. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell 75:1305-1315. [DOI] [PubMed] [Google Scholar]
- 17.Fingar, D. C., and J. Blenis. 2004. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23:3151-3171. [DOI] [PubMed] [Google Scholar]
- 18.Gao, X., Y. Zhang, P. Arrazola, O. Hino, T. Kobayashi, R. S. Yeung, B. Ru, and D. Pan. 2002. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat. Cell Biol. 4:699-704. [DOI] [PubMed] [Google Scholar]
- 19.Garami, A., F. J. Zwartkruis, T. Nobukuni, M. Joaquin, M. Roccio, H. Stocker, S. C. Kozma, E. Hafen, J. L. Bos, and G. Thomas. 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell 11:1457-1466. [DOI] [PubMed] [Google Scholar]
- 20.Goncharova, E. A., D. A. Goncharov, A. Eszterhas, D. S. Hunter, M. K. Glassberg, R. S. Yeung, C. L. Walker, D. Noonan, D. J. Kwiatkowski, M. M. Chou, R. A. Panettieri, Jr., and V. P. Krymskaya. 2002. Tuberin regulates p70 S6 kinase activation and ribosomal protein S6 phosphorylation. A role for the TSC2 tumor suppressor gene in pulmonary lymphangioleiomyomatosis (LAM). J. Biol. Chem. 277:30958-30967. [DOI] [PubMed] [Google Scholar]
- 21.Im, E., F. C. von Lintig, J. Chen, S. Zhuang, W. Qui, S. Chowdhury, P. F. Worley, G. R. Boss, and R. B. Pilz. 2002. Rheb is in a high activation state and inhibits B-Raf kinase in mammalian cells. Oncogene 21:6356-6365. [DOI] [PubMed] [Google Scholar]
- 22.Inoki, K., Y. Li, T. Xu, and K. L. Guan. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17:1829-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoki, K., Y. Li, T. Zhu, J. Wu, and K. L. Guan. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4:648-657. [DOI] [PubMed] [Google Scholar]
- 24.Jacinto, E., and M. N. Hall. 2003. Tor signalling in bugs, brain and brawn. Nat. Rev. Mol. Cell Biol. 4:117-126. [DOI] [PubMed] [Google Scholar]
- 25.Jones, A. C., M. M. Shyamsundar, M. W. Thomas, J. Maynard, S. Idziaszczyk, S. Tomkins, J. R. Sampson, and J. P. Cheadle. 1999. Comprehensive mutation analysis of TSC1 and TSC2—and phenotypic correlations in 150 families with tuberous sclerosis. Am. J. Hum. Genet. 64:1305-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenerson, H. L., L. D. Aicher, L. D. True, and R. S. Yeung. 2002. Activated mammalian target of rapamycin pathway in the pathogenesis of tuberous sclerosis complex renal tumors. Cancer Res. 62:5645-5650. [PubMed] [Google Scholar]
- 27.Krymskaya, V. P. 2003. Tumour suppressors hamartin and tuberin: intracellular signalling. Cell. Signal. 15:729-739. [DOI] [PubMed] [Google Scholar]
- 28.Kwiatkowski, D. J. 2003. Tuberous sclerosis: from tubers to mTOR. Ann. Hum. Genet. 67:87-96. [DOI] [PubMed] [Google Scholar]
- 29.Kwiatkowski, D. J., H. Zhang, J. L. Bandura, K. M. Heiberger, M. Glogauer, N. el-Hashemite, and H. Onda. 2002. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 11:525-534. [DOI] [PubMed] [Google Scholar]
- 30.Li, Y., M. N. Corradetti, K. Inoki, and K. L. Guan. 2004. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem. Sci. 29:32-38. [DOI] [PubMed] [Google Scholar]
- 31.Li, Y., K. Inoki, R. Yeung, and K. L. Guan. 2002. Regulation of TSC2 by 14-3-3 binding. J. Biol. Chem. 277:44593-44596. [DOI] [PubMed] [Google Scholar]
- 32.Mach, K. E., K. A. Furge, and C. F. Albright. 2000. Loss of Rhb1, a Rheb-related GTPase in fission yeast, causes growth arrest with a terminal phenotype similar to that caused by nitrogen starvation. Genetics 155:611-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maheshwar, M. M., J. P. Cheadle, A. C. Jones, J. Myring, A. E. Fryer, P. C. Harris, and J. R. Sampson. 1997. The GAP-related domain of tuberin, the product of the TSC2 gene, is a target for missense mutations in tuberous sclerosis. Hum. Mol. Genet. 6:1991-1996. [DOI] [PubMed] [Google Scholar]
- 34.Maheshwar, M. M., R. Sandford, M. Nellist, J. P. Cheadle, B. Sgotto, M. Vaudin, and J. R. Sampson. 1996. Comparative analysis and genomic structure of the tuberous sclerosis 2 (TSC2) gene in human and pufferfish. Hum. Mol. Genet. 5:131-137. [DOI] [PubMed] [Google Scholar]
- 35.Manning, B. D., and L. C. Cantley. 2003. Rheb fills a GAP between TSC and TOR. Trends Biochem. Sci. 28:573-576. [DOI] [PubMed] [Google Scholar]
- 36.Manning, B. D., A. R. Tee, M. N. Logsdon, J. Blenis, and L. C. Cantley. 2002. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/Akt pathway. Mol. Cell 10:151-162. [DOI] [PubMed] [Google Scholar]
- 37.Maruta, H., J. Holden, A. Sizeland, and G. D'Abaco. 1991. The residues of Ras and Rap proteins that determine their GAP specificities. J. Biol. Chem. 266:11661-11668. [PubMed] [Google Scholar]
- 38.Marygold, S. J., and S. J. Leevers. 2002. Growth signaling: TSC takes its place. Curr. Biol. 12:R785-R787. [DOI] [PubMed] [Google Scholar]
- 39.Nellist, M., M. A. van Slegtenhorst, M. Goedbloed, A. M. van den Ouweland, D. J. Halley, and P. van der Sluijs. 1999. Characterization of the cytosolic tuberin-hamartin complex. Tuberin is a cytosolic chaperone for hamartin. J. Biol. Chem. 274:35647-35652. [DOI] [PubMed] [Google Scholar]
- 40.Onda, H., P. B. Crino, H. Zhang, R. D. Murphey, L. Rastelli, B. E. G. Rothberg, and D. J. Kwiatkowski. 2002. Tsc2 null murine neuroepithelial cells are a model for human tuber giant cells, and show activation of an mTOR pathway. Mol. Cell. Neurosci. 21:561-574. [DOI] [PubMed] [Google Scholar]
- 41.Patel, P. H., N. Thapar, L. Guo, M. Martinez, J. Maris, C. L. Gau, J. A. Lengyel, and F. Tamanoi. 2003. Drosophila Rheb GTPase is required for cell cycle progression and cell growth. J. Cell Sci. 116:3601-3610. [DOI] [PubMed] [Google Scholar]
- 42.Plank, T. L., R. S. Yeung, and E. P. Henske. 1998. Hamartin, the product of the tuberous sclerosis 1 (TSC1) gene, interacts with tuberin and appears to be localized to cytoplasmic vesicles. Cancer Res. 58:4766-4770. [PubMed] [Google Scholar]
- 43.Porfiri, E., T. Evans, P. Chardin, and J. F. Hancock. 1994. Prenylation of Ras proteins is required for efficient hSOS1-promoted guanine nucleotide exchange. J. Biol. Chem. 269:22672-22677. [PubMed] [Google Scholar]
- 44.Proud, C. G. 2002. Regulation of mammalian translation factors by nutrients. Eur. J. Biochem. 269:5338-5349. [DOI] [PubMed] [Google Scholar]
- 45.Saucedo, L. J., X. Gao, D. A. Chiarelli, L. Li, D. Pan, and B. A. Edgar. 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5:566-571. [DOI] [PubMed] [Google Scholar]
- 46.Scheffzek, K., M. R. Ahmadian, W. Kabsch, L. Wiesmuller, A. Lautwein, F. Schmitz, and A. Wittinghofer. 1997. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277:333-338. [DOI] [PubMed] [Google Scholar]
- 47.Scheffzek, K., M. R. Ahmadian, and A. Wittinghofer. 1998. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem. Sci. 23:257-262. [DOI] [PubMed] [Google Scholar]
- 48.Seeburg, P. H., W. W. Colby, D. J. Capon, D. V. Goeddel, and A. D. Levinson. 1984. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature 312:71-75. [DOI] [PubMed] [Google Scholar]
- 49.Stewart, S., and K. L. Guan. 2000. The dominant negative Ras mutant, N17Ras, can inhibit signaling independently of blocking Ras activation. J. Biol. Chem. 275:8854-8862. [DOI] [PubMed] [Google Scholar]
- 50.Stocker, H., T. Radimerski, B. Schindelholz, F. Wittwer, P. Belawat, P. Daram, S. Breuer, G. Thomas, and E. Hafen. 2003. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 5:559-565. [DOI] [PubMed] [Google Scholar]
- 51.Tabancay, A. P., Jr., C. L. Gau, I. M. Machado, E. J. Uhlmann, D. H. Gutmann, L. Guo, and F. Tamanoi. 2003. Identification of dominant negative mutants of Rheb GTPase and their use to implicate the involvement of human Rheb in the activation of p70S6K. J. Biol. Chem. 278:39921-39930. [DOI] [PubMed] [Google Scholar]
- 52.Tee, A. R., D. C. Fingar, B. D. Manning, D. J. Kwiatkowski, L. C. Cantley, and J. Blenis. 2002. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. USA 99:13571-13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tee, A. R., B. D. Manning, P. P. Roux, L. C. Cantley, and J. Blenis. 2003. Tuberous sclerosis complex gene products, tuberin and hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13:1259-1268. [DOI] [PubMed] [Google Scholar]
- 54.Urano, J., C. Ellis, G. J. Clark, and F. Tamanoi. 2001. Characterization of Rheb functions using yeast and mammalian systems. Methods Enzymol. 333:217-231. [DOI] [PubMed] [Google Scholar]
- 55.Yamagata, K., L. K. Sanders, W. E. Kaufmann, W. Yee, C. A. Barnes, D. Nathans, and P. F. Worley. 1994. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J. Biol. Chem. 269:16333-16339. [PubMed] [Google Scholar]
- 56.Yang, W., A. P. Tabancay, Jr., J. Urano, and F. Tamanoi. 2001. Failure to farnesylate Rheb protein contributes to the enrichment of G0/G1 phase cells in the Schizosaccharomyces pombe farnesyltransferase mutant. Mol. Microbiol. 41:1339-1347. [DOI] [PubMed] [Google Scholar]
- 57.Young, J., and S. Povey. 1998. The genetic basis of tuberous sclerosis. Mol. Med. Today 4:313-319. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, F. L., and P. J. Casey. 1996. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65:241-269. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, Y., X. Gao, L. J. Saucedo, B. Ru, B. A. Edgar, and D. Pan. 2003. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5:578-581. [DOI] [PubMed] [Google Scholar]