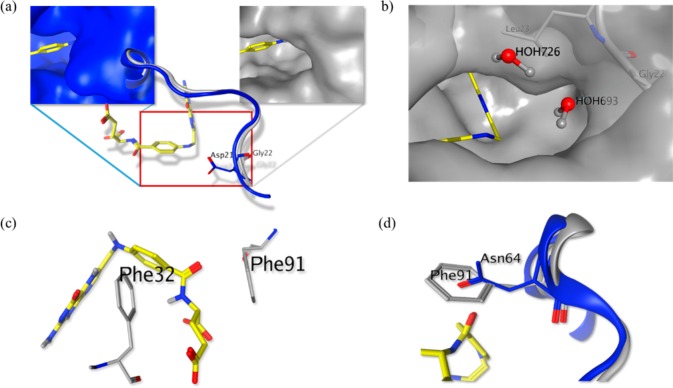
Figure 2.
Structural differences between TgDHFR and hDHFR targeted for rational inhibitor design. (a) Structural comparison of TgDHFR (gray; PDB: 4KYA) and hDHFR (blue; PDB: 4M6K), indicating the presence of a glycine residue (G22) in TgDHFR relative to an aspartic acid (D21) in hDHFR. (b) Presence of a leucine residue (L23) adjacent to G22 creates a solvent-exposed, hydrophobic cavity. Water molecules are shown occupying the cavity as predicted by 3D-RISM (MOE, CCG, Inc.). (c) F32 and F91 in TgDHFR are appropriately positioned for participation in π–π interactions with small molecule ligands. (d) Presence of an asparagine (N64) in hDHFR (blue) at the same position as F91 in TgDHFR (gray) indicates that achieving π–π stacking could modulate species specificity.