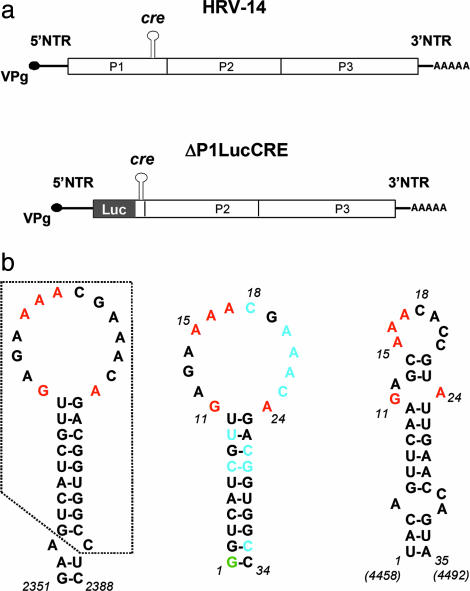
Fig. 1.
Schematic of the HRV-14 genome and replicon and predicted cre structures. (a) (Upper) Organization of the HRV-14 genome showing the position of the critical internally located cre that is required for RNA replication. The polyprotein-coding region is indicated by the open box, with its three major segments P1 (capsid proteins), P2, and P3 (replicase proteins) identified. (Lower) Schematic of the subgenomic HRV-14 ΔP1LucCRE replicon (2) used for replication studies. Most of the capsid protein sequence is replaced with the luciferase (Luc) coding sequence, and replication is monitored by assaying for luciferase after transfection of HeLa cells. (b) Nucleotide sequences and mfold-predicted secondary structures of the HRV-14 and poliovirus cres. (Left) Predicted secondary RNA structure in the region spanning nucleotides 2351-2388 within the P1 segment of the HRV-14 genome (2). The minimal functional cre (10) comprises the boxed sequence. Nucleotides within the cre loop that have been shown by mutational analysis to be critical for VPg uridylylation and RNA replication are shown in red (10). (Center) Predicted secondary structure of RNA transcripts subjected to NMR analysis (Δg = -8.8 kcal/mol). A 5′ terminal G (green) was added to the minimal cre sequence to facilitate RNA synthesis by the T7 polymerase; the two G:C base pairs at the end of the stem also reduce the possibility of end-fraying. Residues shown in blue are those that adopt C2-endo sugar puckering in the structural model. (Right) Predicted secondary structure of the functionally related cre located between nucleotides 4458 and 4492 within the P2 segment of the poliovirus genome (Δg = -5.0 kcal/mol). Conserved purine residues are highlighted in red.