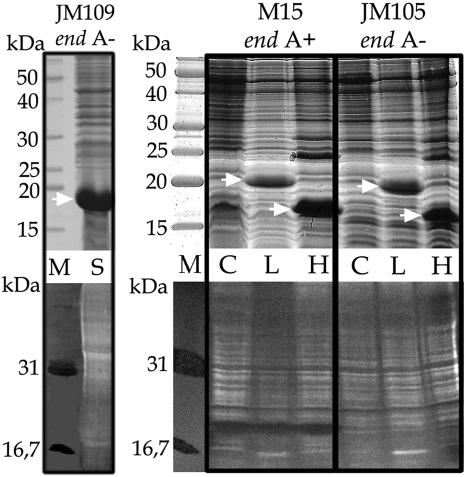
FIG. 1.
Recombinant L. pneumophila LpCyp18, human CypA, and S. antibioticus SanCyp18 cyclophilins were expressed in different E. coli host strains (strain M15, endA+; strains JM109 and JM105, endA mutant). Frozen (−70°C) cells were thawed, resuspended in 15 ml of buffer (20 mM Tris-HCl [pH 8.8], 1 mM EDTA, 7 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride), and ruptured on ice in an MSE Soniprep 150 ultrasonic disintegrator for 4 cycles of 10 s. After centrifugation at 10,000 rpm in an Eppendorf 5415 R microcentrifuge for 30 min at 4°C, the supernatant was used to analyze protein and nuclease activity. Upper panels, Coomassie-stained proteins (50 μg per lane) analyzed by SDS-PAGE; lower panels, activity gel analysis of the corresponding samples. The strains of E. coli used are indicated at the top of the panels. Lanes M, molecular mass markers (in kilodaltons) (upper panel) and activity gel markers (DNase I, 31 kDa; micrococcal nuclease, 16.7 kDa) (lower panel); lanes C, control E. coli extract without expression vector; lanes L, extract with overexpressed LpCyp18; lanes H, extract with overexpressed human CypA; lane S, extract with overexpressed SanCyp18. Activity is detected as a black band within the brighter background. The white bands along the lanes represent the fragmented chromosomal DNA of E. coli. Arrows show the positions of the recombinant cyclophilins.