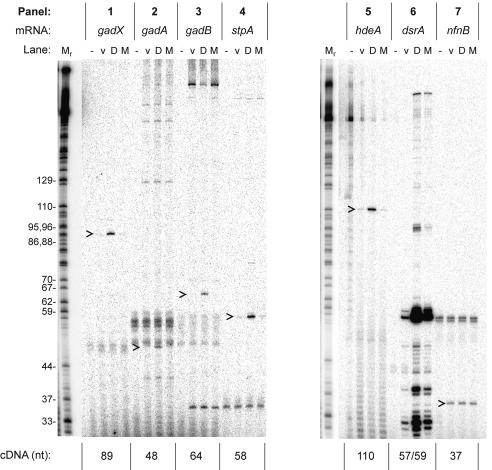
FIG. 1.
Quantitative primer extension analysis of acid resistance genes. RNA was extracted from cells with a vector plasmid (lanes v), a DsrA-overproducing plasmid (lanes D), or a plasmid overproducing inactive mutant DsrA (lanes M). A minus sign indicates a no-RNA control lane. The labeled cDNA products of primer extension were analyzed by polyacrylamide gel electrophoresis; representative gel data are shown. The size of the major cDNA product is given in nucleotides below the panel. The name of each mRNA (top) corresponds to transcripts originating from the major promoter of each gene. The nfnB gene was tested as an unregulated control. Carets indicate relevant primer extension products. A dideoxy-GTP sequencing ladder of an unrelated RNA (Tetrahymena thermophila L-21 IVS) was used as a size marker. The values on the left are sizes in nucleotides. nt, nucleotides.