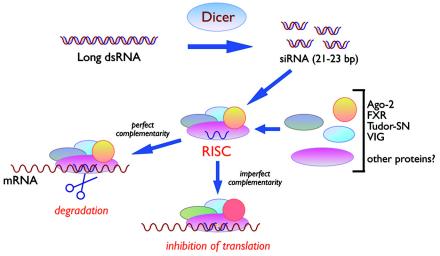
FIG. 3.
Mechanism of RNAi. In mammalian cells, long dsRNAs are trimmed in the nucleus by Dicer to siRNAs of 21 to 23 bp, which are then assembled into RISCs. During or after the assembly process, the two siRNA strands are unwound, and only one remains in active RISCs. These recognize their cytoplasmic mRNA targets by complementarity base pairing and direct mRNA degradation. In the case of micro-RNAs, nuclear precursors are first trimmed by the Drosha enzyme into highly structured RNAs of about 70 nucleotides in length. In the cytoplasm, these are further processed by Dicer to yield mature micro-RNAs that assemble into complexes related to RISCs. In this case, target sequences in mRNA 3′ untranslated regions are recognized by imperfect base pairing (indicated as a yellow bar in the siRNA-mRNA hybrid at the bottom) and lead to translation inhibition by a still-unclear mechanism. See the text for details.