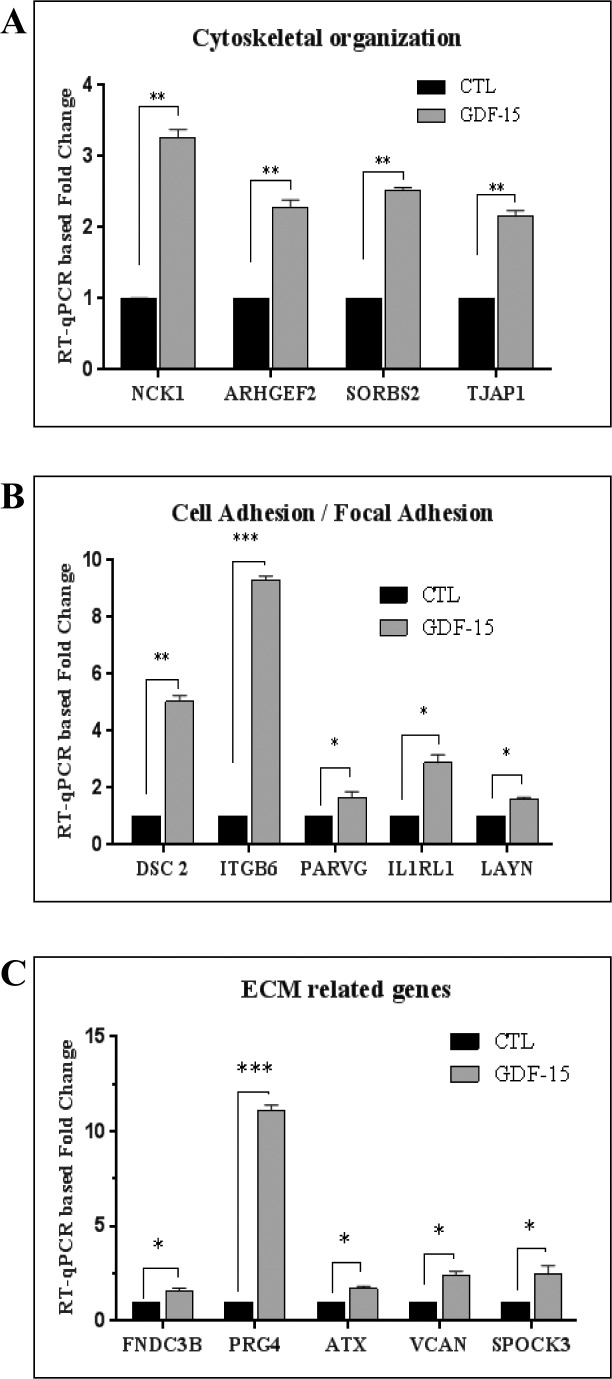
Figure 4.
Real-time qPCR–based confirmation of GDF-15–induced gene expression in human TM cells. An RT-qPCR–based approach was undertaken to independently validate the data obtained from cDNA microarray analyses of the effects of GDF-15 on gene expression in TM cells. A portion of the RNA submitted for cDNA microarray analysis was reverse transcribed and used for the RT-qPCR experiment along with another biologic replicate of TM cells derived from a 71-year-old donor. The selected genes were categorized based on their Gene Ontology terms and clustered into three different groups namely; (A) Cytoskeletal organization (NCK1, ARHGEF2, SORBS2, and TJAP1), (B) Cell adhesion (DSC2, ITGB6, PARVG, IL1RL1, and LAYN), and (C) ECM-related genes (FNDC3B, PRG4, ATX, SPOCK3, and VCAN). Real-time quantification of the expression of selected genes was normalized to the cycle values of GAPDH. Fold changes were calculated based on the values from triplicate analyses of two individual samples (n = 6, mean ± SEM). The selected genes showed a significant increase (in fold change) in expression upon GDF-15 treatment as compared to corresponding controls. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.