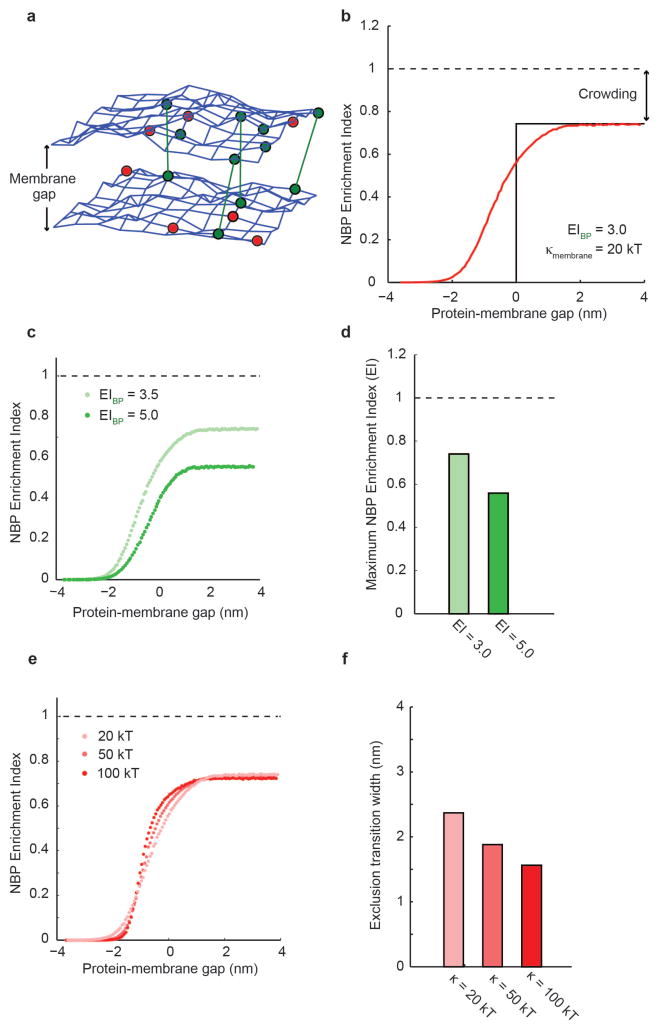
Figure 5.
Monte Carlo simulations of size-dependent protein segregation. (a) A fluctuating triangulated mesh represents each of the two membrane surfaces that form a membrane interface in the simulations. Nodes of the mesh can be occupied by non-binding proteins (red) and by binding proteins (green) that may form bonds across the interface (green lines). (b) Simulation of protein enrichment for a non-binding protein as a function of the protein-membrane gap. At positive protein-membrane gap (right), non-binding proteins are smaller than the membrane interface, and lateral crowding in the plane of the membrane results in a maximum enrichment index, or maximum inclusion, below 1.0. At negative protein-membrane gap (left), non-binding proteins are completely excluded to minimize bending of the membrane. Near a protein-membrane gap of 0, fluctuations of the membrane surface and steric interactions between apposite membranes and proteins result in a smooth transition between these two extremes. (c & d) The maximum enrichment index, or maximum inclusion, at large protein-membrane gaps decreases with increasing binding protein enrichment, since increased binding protein density within the interface crowds out non-binding proteins. (e & f) Increasing the bending rigidity of the membrane makes the transition between maximum inclusion and maximum exclusion of non-binding proteins sharper. If the exclusion transition width is defined as the width along the protein-membrane gap axis between 10 and 90 of maximum inclusion, the change in transition width is less than 1 nm for a factor of five increase in bending rigidity, suggesting that size-dependent segregation is only modestly affected by bending rigidity.