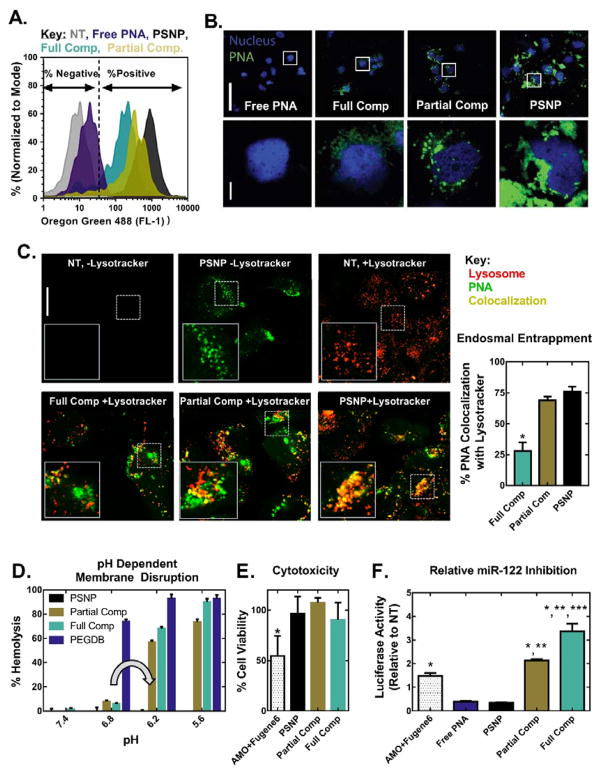
Figure 2.
Coating of PSNPs with PEGDB decreases PNA uptake but increases both endosome escape potential and anti-miRNA activity relative to uncoated PSNPs in Huh7 human hepatocellular carcinoma cells. PEGDB functionalization decreases cellular uptake, as characterized by (A) flow cytometry and (B) confocal microscopy, 24 h after treatment with Alexa Fluor 488–labeled anti-miR122 PNA at a 2 μM PNA dose. Top scale bar = 100 μm, bottom scale bar = 10 μm. (C) PEGDB functionalization increases PNA cytosolic delivery, as shown by colocalization analysis of Alexa Fluor 488–labeled PNA with LysoTracker at 24 h after treatment with 2 μM PNA. Endosomal entrapment was quantified by calculating the Manders’ overlap coefficients for green and red pixels, shown at the right as means ± SEM (n ≥ 3 separate images). Increased cytosolic delivery observed for composite particles is due to (D) the pH dependent membrane disruptive function (grey arrow) of PEGDB, as determined by a hemolysis assay. Composites did not disrupt erythrocyte membranes at pH 7.4, but produced robust hemolysis at pH 6.2, which is representative of late endosomes. (E) A firefly luciferase assay reveals that all PSNP treatments are non-toxic at a 2 μM PNA dose, in contrast to the gold-standard of a 2’OMe modified RNA delivered using a commercial cationic transfection reagent (AMO+Fugene6). (F) Therapeutic anti-miR122 activity increases with increasing PEGDB polymer functionalization (based on renilla luciferase readout tied directly to miR-122 inhibition) 24 hours after treatment, when compared to free, unencapsulated PNA and the control, 2’OMe AMO. (p<0.05 when compared to *Free PNA or PSNP, **AMO+Fugene6, and ***Partial Comp).