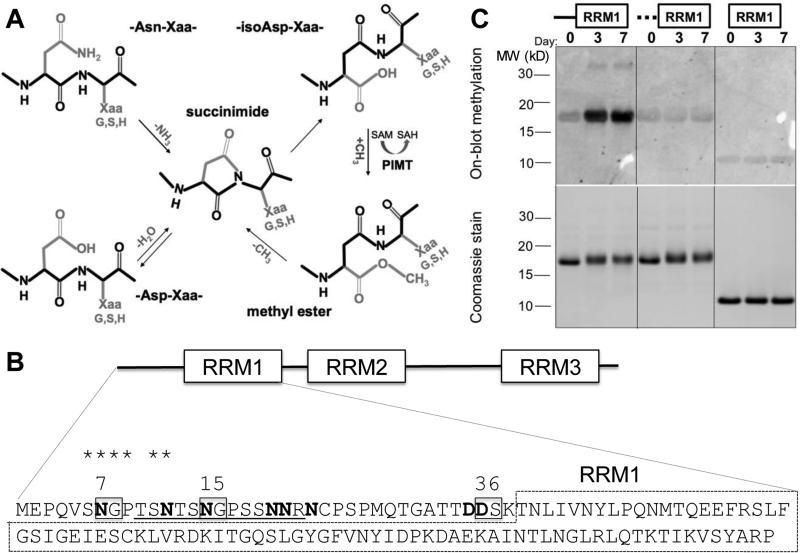
Fig. 1.
ELAVL4 N-terminal region becomes isoaspartylated in vitro. (A) Mechanism of isoAsp formation and repair. Isoaspartylation occurs by spontaneous dehydration or deamidation of an asparagine (Asn-Xaa) or aspartate (Asp-Xaa) linkage (where Xaa is usually glycine, serine or histidine). This produces a metastable succinimide intermediate, which in turn undergoes spontaneous hydrolysis to generate an unequal mixture, typically 15–30% of the normal L-aspartyl linkage and 70–85% of an abnormal L-isoaspartyl-Xaa linkage with a kinked protein backbone (protein backbone indicated in black). PIMT converts L-isoaspartyl sites to α-carboxyl-O-methyl esters, converting methyl donor S-adenosyl methionine (SAM) to S-adenosyl homocysteine (SAH). At physiological pH and temperature, the α-carboxyl-O-methyl esters have a typical half-life of a few minutes and undergo spontaneous demethylation to reform the succinimide intermediate. Each PIMT cycle typically repairs ~25% of the isoAsp peptide bond by converting it to a normal aspartyl peptide. Unrepaired peptides are continuously recycled, and the overall repair efficiency is 85% or greater. (B) Canonical isoAsp sites (N or D followed by S, G or H in unstructured regions) are boxed, all N and D with potential for isoaspartylation are bolded. It is unlikely that the globular structure of the RRM1 domain (dashed box) allows isoaspartylation of RRM1 residues. The isoAsp-containing peptide used to immunize rabbits is underlined and homologous amino acids shared between this isoAsp-containing peptide and the peptide sequence surrounding N7 are starred; (C) Recombinant wild type ELAVL41-117 protein, a mutant with substitutions at all N (to Q) and D (to E) residues in the N-terminal 38 amino acids (indicated by the dashed line), and ELAVL439-117, a protein without the N-terminal region, were incubated at physiological conditions for 0, 3, and 7 days, separated by gel electrophoresis, transferred to a membrane and subjected to on-blot labeling with PIMT and 3H-SAM (top panel). Only wild type ELAVL41-117 showed above background labeling by PIMT at 3 and 7 days. The bottom panel shows a Coomassie-stained gel, indicating similar loading of wild type, mutant, and truncated proteins over the incubation period. A representative image of several similar experiments is shown.