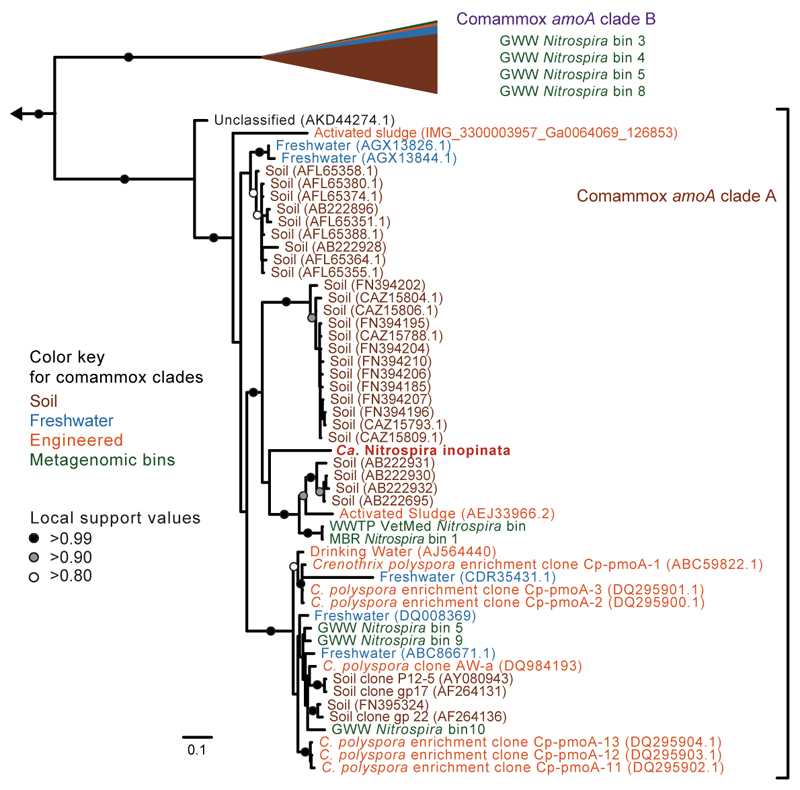
Extended Data Figure 8. Phylogenetic affiliation of comammox amoA sequences to amoA sequences from different environments.
Bayesian inference tree showing the phylogenetic relationship of the amoA sequences from Ca. N. inopinata and metagenomic bins from this study (224 taxa, 939 nucleotide alignment positions). Ca. N. inopinata clusters confidently into comammox amoA clade A. Comammox amoA clade B (116 taxa) has been collapsed for clarity and the proportion of database sequences from soil (95 taxa), freshwater (13 taxa), and engineered environments (4 taxa) is represented as a proportion of the collapsed clade. AmoA from the metagenomic Nitrospira bins generated for this study (5 taxa in clade A, 4 taxa in clade B) are numbered as in Supplementary Table 8. Scale bar indicates estimated change per nucleotide. The outgroup consists of 27 betaproteobacterial amoA and 29 diverse pmoA sequences.