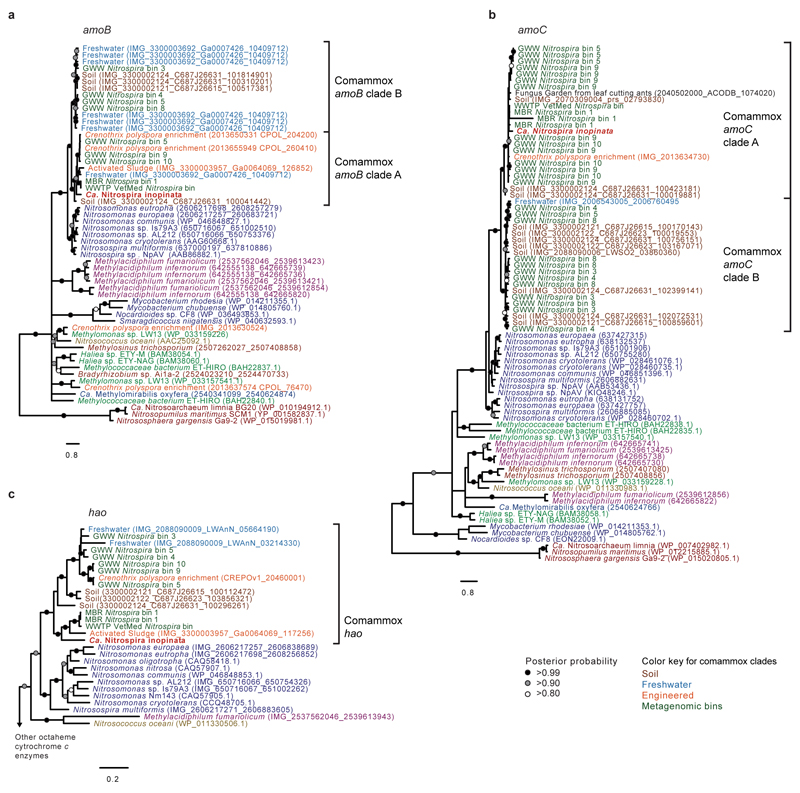
Extended Data Figure 9. Phylogenetic relationship of comammox amoB, amoC, and hao sequences to corresponding gene family members.
Trees were calculated with PhyloBayes using nucleotide sequences aligned according to their amino acid translations. Support values indicate the consensus probability from 5 independent chains. Sequences outside the comammox clades are colored as in main text Figure 3. Metagenomic bins are numbered as in Supplementary Table 8. Scale bars indicate the estimated substitutions per nucleotide. a, Phylogenetic relationship of Ca. N. inopinata amoB to other amoB and pmoB genes (57 taxa, 1,518 alignment positions). b, Phylogenetic relationship of Ca. N. inopinata amoC to other amoC and pmoC genes (81 taxa, 993 alignment positions). c, Phylogenetic relationship of Ca. N. inopinata hydroxylamine dehydrogenase (hao) to other hao genes (37 taxa, 2,875 alignment positions).