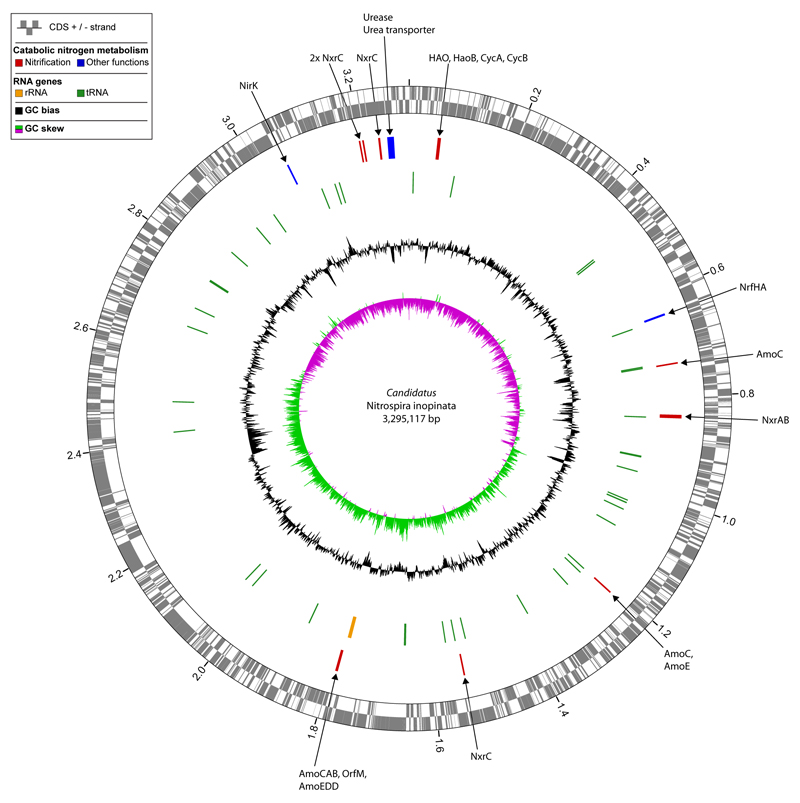
Extended Data Figure 3. Circular representation of the Ca. N. inopinata chromosome.
Predicted coding sequences (CDS; rings 1+2), genes of enzymes involved in nitrification and other pathways of catabolic nitrogen metabolism (ring 3), RNA genes (ring 4), and local nucleotide composition measures (rings 5+6) are shown. Very short features were enlarged to enhance visibility. Clustered genes, such as several tRNA genes, may appear as one line due to space limitations. The tick interval is 0.2 Mbp. Abbreviations: Amo, ammonia monooxygenase; HAO, hydroxylamine dehydrogenase; CycA and CycB, tetraheme c-type cytochromes that form the hydroxylamine ubiquinone redox module together with HAO; NirK, Cu-dependent nitrite reductase; Nrf, cytochrome c nitrite reductase; Nxr, nitrite oxidoreductase; Orf, open reading frame.