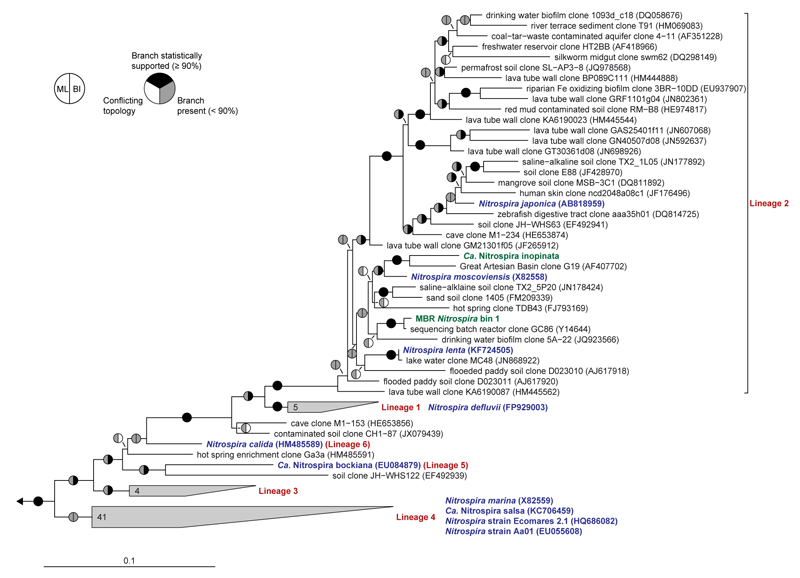
Extended Data Figure 4. Phylogenetic affiliation of Ca. N. inopinata.
The maximum likelihood tree, which is based on 16S ribosomal RNA sequences of cultured and uncultured representative members of the genus Nitrospira, shows that the comammox organism Ca. N. inopinata (highlighted green) is a member of Nitrospira lineage II. Another 16S rRNA gene sequence was extracted from MBR metagenomic Nitrospira bin 1 (also highlighted green). This sequence bin also contained amo and hao genes (main text Figure 1, Extended Data Figures 8 and 9). The cultured Nitrospira strains other than Ca. N. inopinata, which are not known to use ammonia as a source of energy and reductant, are highlighted blue. Nitrospira lineages are labeled red. Pie charts indicate statistical support of branches based on maximum likelihood (ML; 1,000 bootstrap iterations) and Bayesian inference (BI; posterior probability, 4 independent chains). In total, 95 taxa and 1,543 nucleotide sequence alignment positions were considered. Numbers in wedges indicate the numbers of taxa. The scale bar indicates 0.1 estimated substitutions per nucleotide.