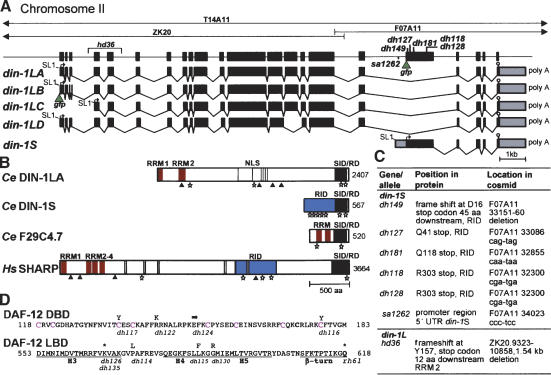
Figure 2.
din-1 structure. (A) Cosmids from the din-1 region, gene structure with mutations and din-1S::gfp insertion site (green triangle) are shown. (Bottom) mRNA isoforms containing SL1 trans-splice leader (line), start codon (arrow), stop codon (stop sign), and polyadenylation site (poly A). gfp insertion site of din-1LB is shown. (B) Domain structure of DIN-1 and homologs. (RRM) RNA recognition motif (red); (NLS) Nuclear localization sites (gray); (RID) nuclear receptor interaction domain (blue); (SID/RD) SMRT interaction domain/repression domain (black); (stars) L/IXXL/I/VL/I motifs; (triangles) AKT sites. (C) din-1 mutations. (D) daf-12(rh61) intragenic revertants. (Top) DAF-12 DBD, with conserved cysteines (magenta). (Bottom) LBD helices (H3–5). Amino acid changes are shown above the sequence, allele names below. (Arrow) Splice mutation; (asterisk) stop codon.