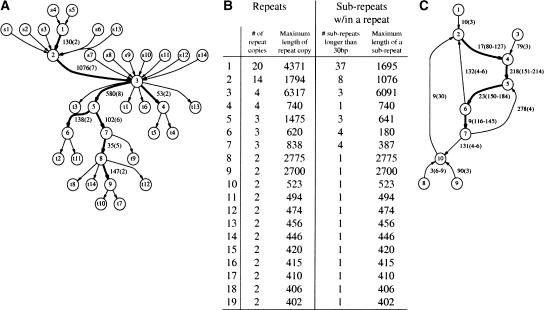
Figure 4.
RepeatGluer representation of a 14-copy transposase IS30 repeat family in the N. meningitidis genome as a mosaic of eight sub-repeats >30 bp (shown by bold edges). The similarity matrix A is constructed based on a 90% similarity threshold and minimum repeat length 400. (A) Transposase IS30 family tangle (with adjacent edges) in the repeat graph. Every repeat copy corresponds to a path from a source sI to a sink tI. Edge label 130(2) indicates the length of the sub-repeat (130 nt) and its multiplicity (2). (B) Summary of all 19 repeat families (>400 bp with 90% similarity threshold) produced by RepeatGluer in the N. meningitidis genome. (C) Simplified tangle corresponding to the most common Alu repeat family in the first 1 Mb of human Chromosome X. Because the real tangle (even for 1 Mb sequence, let alone the entire genome) is too complicated, we removed all edges with multiplicities smaller than 3. This Alu repeat is broken into four sub-repeats with multiplicities ranging from 116 to 214. The multiplicities of every sub-repeat vary along the length of sub-repeat because many Alu repeats are incomplete versions of the canonical Alu (e.g., the multiplicity of the sub-repeat of length 218 varies from 151 to 214).