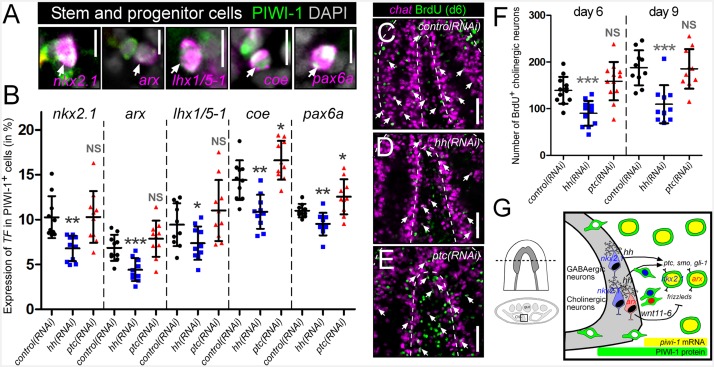
Figure 6. hh signaling regulates homeostatic neurogenesis.
(A) Single confocal plane images displaying examples of neural progenitor cells (white arrow), PIWI-1+ stem/progenitor cells that express neural transcription factors. Scale bar = 10 µM. (B) Quantification of neural progenitor levels in animals with altered levels of hh signaling. Graphs are dot plots measuring the percentage of PIWI-1+ cells that express each neural transcription factor. n = 10, *p<0.05, **p<0.01, ***p<0.001, error bars are standard deviation. (C–E) Single confocal planes displaying newly generated cholinergic neurons in the planarian brain 6 days after a single BrdU pulse (white arrows). Scale bar = 100 µm. (F) Dot plot quantifying the number of new cholinergic neurons. n ≥ 10, ***p<0.001, error bars are standard deviation. (G) Model displaying nkx2.1+ and arx+ neural stem and progenitor cells and the mature neurons that they produce. These same neural cell types also express the hh signaling molecule, which signals back onto adult stem cells, maintaining normal proliferation levels, and homeostatic neurogenesis.