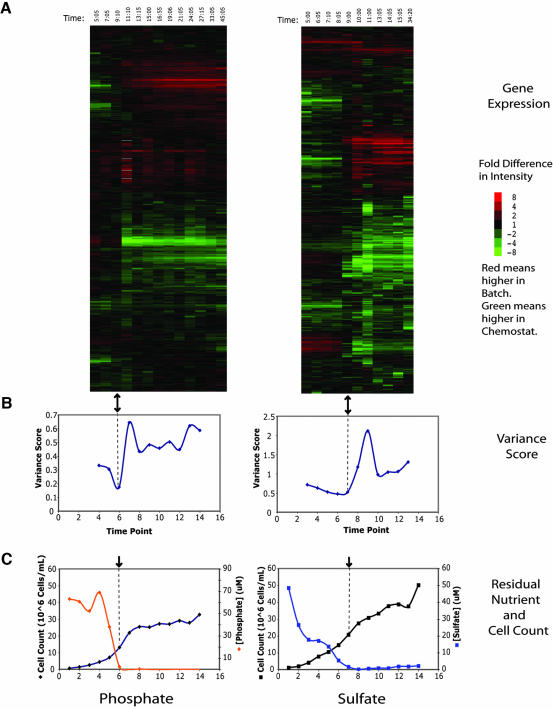
Figure 1.
Global cluster analysis of phosphate and sulfate expression data. (A) Clustergram of genome-wide microarray data for phosphate (P1) and sulfate (S4) time courses. The array indicated by the black arrow has similar representation of transcripts in both channels. It should be noted that the data have not been centered, thus allowing the accurate representation of a constant bias in expression, but also increasing sensitivity to systematic bias. Time points were taken at intervals of approximately 2 h. Each row corresponds to a single gene and each column to a single array. The columns are arranged in order of increasing time. Red values indicate higher expression in the batch, and green values indicate higher expression in the chemostat. The intensity of the color is determined by the fold change and is indicated by the color bar to the right. (B) Variance score has a minimum near the array identified by cluster analysis, shown here with the black arrow. The variance score is a measure of the deviation of the array from equal representation of all transcripts in both channels and is calculated as the average of the square of the log ratios. (C) The limiting nutrient becomes undetectable in the filtered media near the time that the transcriptional state becomes comparable.