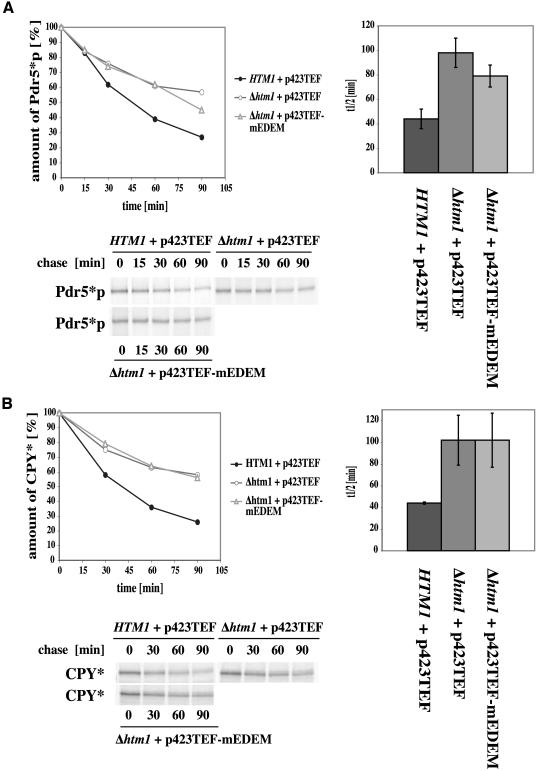
Figure 4.
Influence of mEDEM on glycosylated yeast ERAD substrates in Δhtm1 strains. Pulse-chase analysis was done as described in text. The graphs show average kinetics of three experiments of 90-min pulse-chases and average half-life in wild-type (HTM1 + p423TEF) and Δ htm1 deletion strains with (p423TEF-mEDEM) or without (p423TEF) mouse EDEM. Error bars indicate the variations of the different experiments. The autoradiographs show typical results. (A) Pdr5*p degradation shows a barely significant increase in a strain with mEDEM expression. (B) mEDEM does not show a significant increase of CPY* degradation.