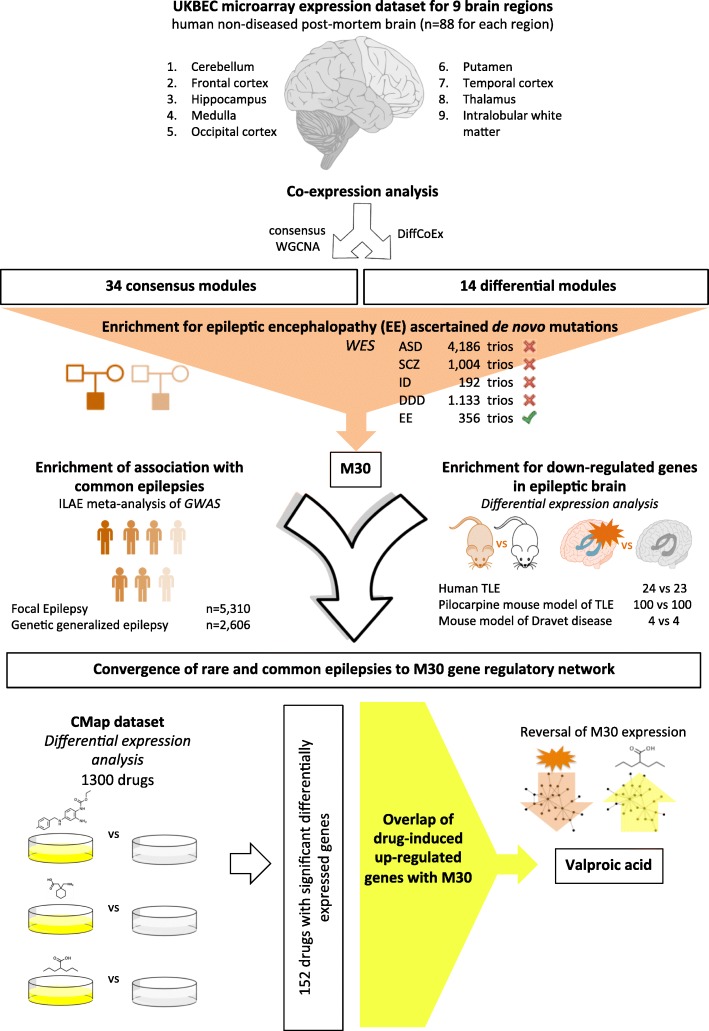
Fig. 1.
Schematic overview of study design. We hypothesised that gene regulatory networks in the human healthy brain disrupted by de novo mutations ascertained from patients with epileptic encephalopathy could be informative for molecular processes involved in different types of epilepsy. We used post-mortem human brain samples ascertained from individuals with no history of psychiatric or neurological illness from the UK Brain Expression Consortium (UKBEC) dataset to build gene co-expression networks (modules) that are expressed across the whole brain (‘consensus’ modules, using the WCGNA consensus method), or differential to one brain region or another (‘differential’ modules, using DiffCoEx method). In order to prioritise modules relevant to epilepsy, we integrated modules with whole-exome-sequencing (WES) studies data of rare de novo mutations ascertained from patients with epileptic encephalopathy (EE) and neurodevelopmental disease more generally (ASD autism spectrum disorder, SCZ schizophrenia, ID intellectual disabilities, DDD broad developmental disease from the Deciphering Developmental Disorders study). A single module was selected in this way: M30. Our hypothesis was that disruption of this gene network might lead to different types of epilepsy. M30 was therefore tested for enrichment of association to common forms of epilepsy using GWAS data from the International League against Epilepsy (ILAE) meta-analysis. Analysis of network genes’ expression in disease in three epilepsies suggested functional disruption and/or downregulation of the network as a common mechanism regulating susceptibility to epilepsy broadly and therefore that the network itself might be targeted as a novel therapeutic strategy. As a proof of concept, we show that among the drugs capable of inducing transcriptional changes in cells of the Connectivity map (Cmap) dataset, VPA, a widely used AED with a broad spectrum of clinical efficacy, is the one that is most significantly predicted to restore the expression of M30 in epilepsy toward health. Full details relating to datasets, experimental methods and references are provided in the manuscript